Genomic Data Has a Diversity Problem, But Global Efforts Are Underway to Fix It

Genetic data sets skew too European, threatening to narrow who will benefit from future advances.
Genomics has begun its golden age. Just 20 years ago, sequencing a single genome cost nearly $3 billion and took over a decade. Today, the same feat can be achieved for a few hundred dollars and the better part of a day . Suddenly, the prospect of sequencing not just individuals, but whole populations, has become feasible.
The genetic differences between humans may seem meager, only around 0.1 percent of the genome on average, but this variation can have profound effects on an individual's risk of disease, responsiveness to medication, and even the dosage level that would work best.
Already, initiatives like the U.K.'s 100,000 Genomes Project - now expanding to 1 million genomes - and other similarly massive sequencing projects in Iceland and the U.S., have begun collecting population-scale data in order to capture and study this variation.
The resulting data sets are immensely valuable to researchers and drug developers working to design new 'precision' medicines and diagnostics, and to gain insights that may benefit patients. Yet, because the majority of this data comes from developed countries with well-established scientific and medical infrastructure, the data collected so far is heavily biased towards Western populations with largely European ancestry.
This presents a startling and fast-emerging problem: groups that are under-represented in these datasets are likely to benefit less from the new wave of therapeutics, diagnostics, and insights, simply because they were tailored for the genetic profiles of people with European ancestry.
We may indeed be approaching a golden age of genomics-enabled precision medicine. But if the data bias persists then there is a risk, as with most golden ages throughout history, that the benefits will not be equally accessible to all, and existing inequalities will only be exacerbated.
To remedy the situation, a number of initiatives have sprung up to sequence genomes of under-represented groups, adding them to the datasets and ensuring that they too will benefit from the rapidly unfolding genomic revolution.
Global Gene Corp
The idea behind Global Gene Corp was born eight years ago in Harvard when Sumit Jamuar, co-founder and CEO, met up with his two other co-founders, both experienced geneticists, for a coffee.
"They were discussing the limitless applications of understanding your genetic code," said Jamuar, a business executive from New Delhi.
"And so, being a technology enthusiast type, I was excited and I turned to them and said hey, this is incredible! Could you sequence me and give me some insights? And they actually just turned around and said no, because it's not going to be useful for you - there's not enough reference for what a good Sumit looks like."
What started as a curiosity-driven conversation on the power of genomics ended with a commitment to tackle one of the field's biggest roadblocks - its lack of global representation.
Jamuar set out to begin with India, which has about 20 percent of the world's population, including over 4000 different ethnicities, but contributes less than 2 percent of genomic data, he told Leaps.org.
Eight years later, Global Gene Corp's sequencing initiative is well underway, and is the largest in the history of the Indian subcontinent. The program is being carried out in collaboration with biotech giant Regeneron, with support from the Indian government, local communities, and the Indian healthcare ecosystem. In August 2020, Global Gene Corp's work was recognized through the $1 million 2020 Roddenberry award for organizations that advance the vision of 'Star Trek' creator Gene Roddenberry to better humanity.
This problem has already begun to manifest itself in, for example, much higher levels of genetic misdiagnosis among non-Europeans tested for their risk of certain diseases, such as hypertrophic cardiomyopathy - an inherited disease of the heart muscle.
Global Gene Corp also focuses on developing and implementing AI and machine learning tools to make sense of the deluge of genomic data. These tools are increasingly used by both industry and academia to guide future research by identifying particularly promising or clinically interesting genetic variants. But if the underlying data is skewed European, then the effectiveness of the computational analysis - along with the future advances and avenues of research that emerge from it - will be skewed towards Europeans too.
This problem has already begun to manifest itself in, for example, much higher levels of genetic misdiagnosis among non-Europeans tested for their risk of certain diseases, such as hypertrophic cardiomyopathy - an inherited disease of the heart muscle. Most of the genetic variants used in these tests were identified as being causal for the disease from studies of European genomes. However, many of these variants differ both in their distribution and clinical significance across populations, leading to many patients of non-European ancestry receiving false-positive test results - as their benign genetic variants were misclassified as pathogenic. Had even a small number of genomes from other ethnicities been included in the initial studies, these misdiagnoses could have been avoided.
"Unless we have a data set which is unbiased and representative, we're never going to achieve the success that we want," Jamuar says.
"When Siri was first launched, she could hardly recognize an accent which was not of a certain type, so if I was trying to speak to Siri, I would have to repeat myself multiple times and try to mimic an accent which wasn't my accent so that she could understand it.
"But over time the voice recognition technology improved tremendously because the training data was expanded to include people of very diverse backgrounds and their accents, so the algorithms were trained to be able to pick that up and it dramatically improved the technology. That's the way we have to think about it - without that good-quality diverse data, we will never be able to achieve the full potential of the computational tools."
While mapping India's rich genetic diversity has been the organization's primary focus so far, they plan, in time, to expand their work to other under-represented groups in Asia, the Middle East, Africa, and Latin America.
"As other like-minded people and partners join the mission, it just accelerates the achievement of what we have set out to do, which is to map out and organize the world's genomic diversity so that we can enable high-quality life and longevity benefits for everyone, everywhere," Jamuar says.
Empowering African Genomics
Africa is the birthplace of our species, and today still retains an inordinate amount of total human genetic diversity. Groups that left Africa and went on to populate the rest of the world, some 50 to 100,000 years ago, were likely small in number and only took a fraction of the total genetic diversity with them. This ancient bottleneck means that no other group in the world can match the level of genetic diversity seen in modern African populations.
Despite Africa's central importance in understanding the history and extent of human genetic diversity, the genomics of African populations remains wildly understudied. Addressing this disparity has become a central focus of the H3Africa Consortium, an initiative formally launched in 2012 with support from the African Academy of Sciences, the U.S. National Institutes of Health, and the UK's Wellcome Trust. Today, H3Africa supports over 50 projects across the continent, on an array of different research areas in genetics relevant to the health and heredity of Africans.
"Africa is the cradle of Humankind. So what that really means is that the populations that are currently living in Africa are among some of the oldest populations on the globe, and we know that the longer populations have had to go through evolutionary phases, the more variation there is in the genomes of people who live presently," says Zane Lombard, a principal investigator at H3Africa and Associate Professor of Human Genetics at the University of the Witwatersrand in Johannesburg, South Africa.
"So for that reason, African populations carry a huge amount of genetic variation and diversity, which is pretty much uncaptured. There's still a lot to learn as far as novel variation is concerned by looking at and studying African genomes."
A recent landmark H3Africa study, led by Lombard and published in Nature in October, sequenced the genomes of over 400 African individuals from 50 ethno-linguistic groups - many of which had never been sampled before.
Despite the relatively modest number of individuals sequenced in the study, over three million previously undescribed genetic variants were found, and complex patterns of ancestral migration were uncovered.
"In some of these ethno-linguistic groups they don't have a word for DNA, so we've had to really think about how to make sure that we communicate the purposes of different studies to participants so that you have true informed consent," says Lombard.
"The objective," she explained, "was to try and fill some of the gaps for many of these populations for which we didn't have any whole genome sequences or any genetic variation data...because if we're thinking about the future of precision medicine, if the patient is a member of a specific group where we don't know a lot about the genomic variation that exists in that group, it makes it really difficult to start thinking about clinical interpretation of their data."
From H3Africa's conception, the consortium's goal has not only been to better represent Africa's staggering genetic diversity in genomic data sets, but also to build Africa's domestic genomics capabilities and empower a new generation of African researchers. By doing so, the hope is that Africans will be able to set their own genomics agenda, and leapfrog to new and better ways of doing the work.
"The training that has happened on the continent and the number of new scientists, new students, and fellows that have come through the process and are now enabled to start their own research groups, to grow their own research in their countries, to be a spokesperson for genomics research in their countries, and to build that political will to do these larger types of sequencing initiatives - that is really a significant outcome from H3Africa as well. Over and above all the science that's coming out," Lombard says.
"What has been created through H3Africa is just this locus of researchers and scientists and bioethicists who have the same goal at heart - to work towards adjusting the data bias and making sure that all global populations are represented in genomics."
A robot server, controlled remotely by a disabled worker, delivers drinks to patrons at the DAWN cafe in Tokyo.
A sleek, four-foot tall white robot glides across a cafe storefront in Tokyo’s Nihonbashi district, holding a two-tiered serving tray full of tea sandwiches and pastries. The cafe’s patrons smile and say thanks as they take the tray—but it’s not the robot they’re thanking. Instead, the patrons are talking to the person controlling the robot—a restaurant employee who operates the avatar from the comfort of their home.
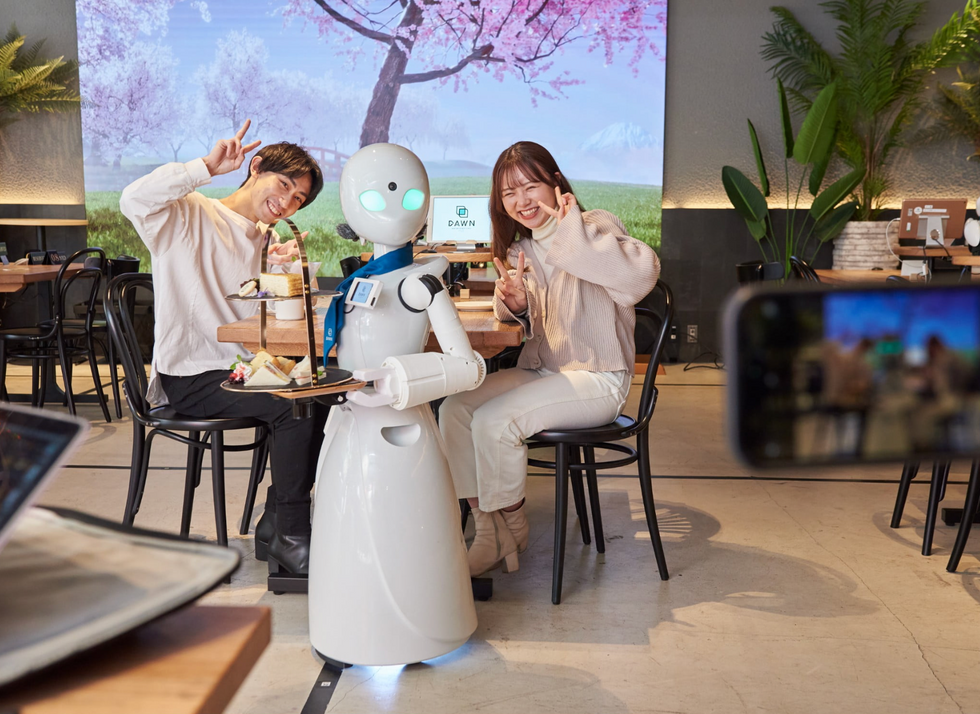
It’s a typical scene at DAWN, short for Diverse Avatar Working Network—a cafe that launched in Tokyo six years ago as an experimental pop-up and quickly became an overnight success. Today, the cafe is a permanent fixture in Nihonbashi, staffing roughly 60 remote workers who control the robots remotely and communicate to customers via a built-in microphone.
More than just a creative idea, however, DAWN is being hailed as a life-changing opportunity. The workers who control the robots remotely (known as “pilots”) all have disabilities that limit their ability to move around freely and travel outside their homes. Worldwide, an estimated 16 percent of the global population lives with a significant disability—and according to the World Health Organization, these disabilities give rise to other problems, such as exclusion from education, unemployment, and poverty.
These are all problems that Kentaro Yoshifuji, founder and CEO of Ory Laboratory, which supplies the robot servers at DAWN, is looking to correct. Yoshifuji, who was bedridden for several years in high school due to an undisclosed health problem, launched the company to help enable people who are house-bound or bedridden to more fully participate in society, as well as end the loneliness, isolation, and feelings of worthlessness that can sometimes go hand-in-hand with being disabled.
“It’s heartbreaking to think that [people with disabilities] feel they are a burden to society, or that they fear their families suffer by caring for them,” said Yoshifuji in an interview in 2020. “We are dedicating ourselves to providing workable, technology-based solutions. That is our purpose.”
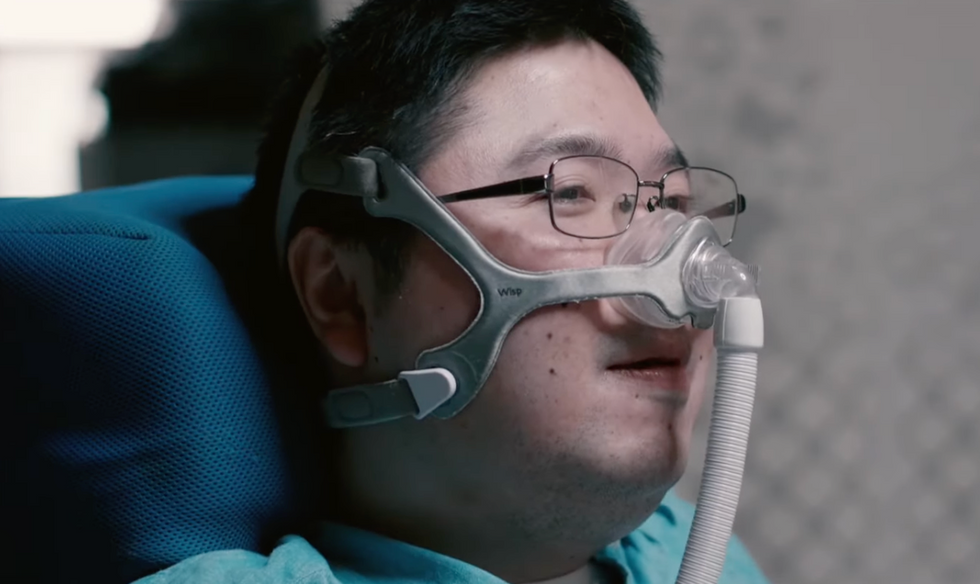
Shota, Kuwahara, a DAWN employee with muscular dystrophy, agrees. "There are many difficulties in my daily life, but I believe my life has a purpose and is not being wasted," he says. "Being useful, able to help other people, even feeling needed by others, is so motivational."
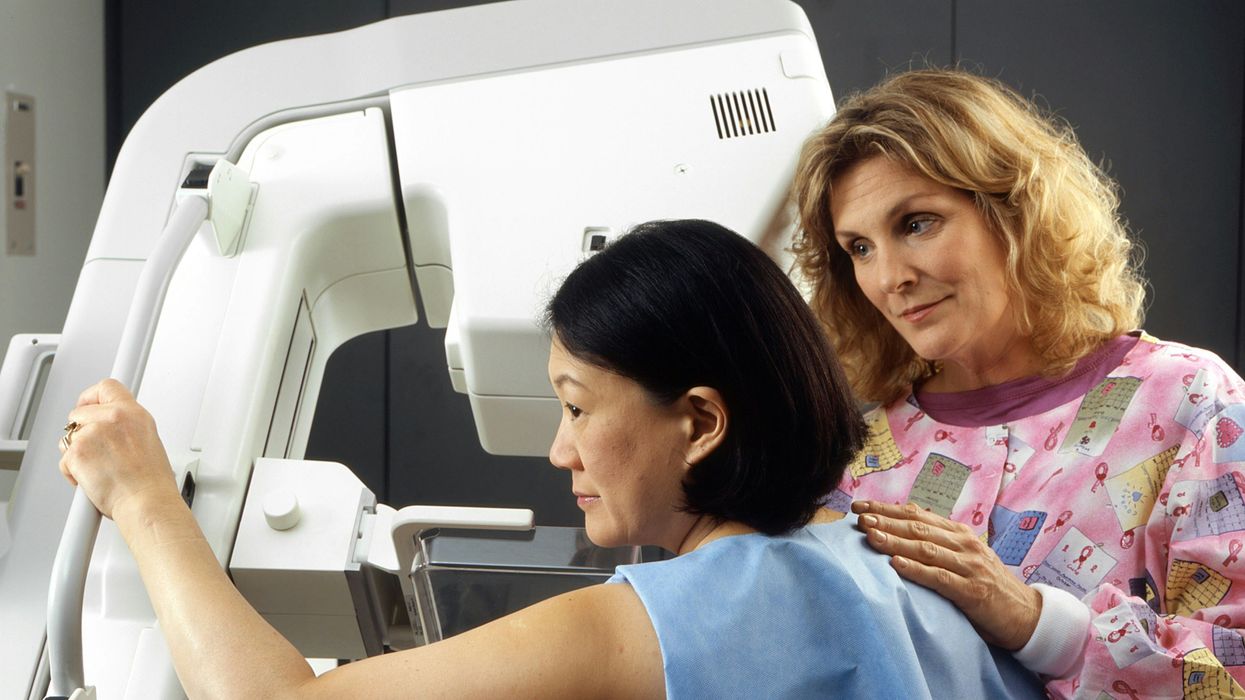
A woman receives a mammogram, which can detect the presence of tumors in a patient's breast.
When a patient is diagnosed with early-stage breast cancer, having surgery to remove the tumor is considered the standard of care. But what happens when a patient can’t have surgery?
Whether it’s due to high blood pressure, advanced age, heart issues, or other reasons, some breast cancer patients don’t qualify for a lumpectomy—one of the most common treatment options for early-stage breast cancer. A lumpectomy surgically removes the tumor while keeping the patient’s breast intact, while a mastectomy removes the entire breast and nearby lymph nodes.
Fortunately, a new technique called cryoablation is now available for breast cancer patients who either aren’t candidates for surgery or don’t feel comfortable undergoing a surgical procedure. With cryoablation, doctors use an ultrasound or CT scan to locate any tumors inside the patient’s breast. They then insert small, needle-like probes into the patient's breast which create an “ice ball” that surrounds the tumor and kills the cancer cells.
Cryoablation has been used for decades to treat cancers of the kidneys and liver—but only in the past few years have doctors been able to use the procedure to treat breast cancer patients. And while clinical trials have shown that cryoablation works for tumors smaller than 1.5 centimeters, a recent clinical trial at Memorial Sloan Kettering Cancer Center in New York has shown that it can work for larger tumors, too.
In this study, doctors performed cryoablation on patients whose tumors were, on average, 2.5 centimeters. The cryoablation procedure lasted for about 30 minutes, and patients were able to go home on the same day following treatment. Doctors then followed up with the patients after 16 months. In the follow-up, doctors found the recurrence rate for tumors after using cryoablation was only 10 percent.
For patients who don’t qualify for surgery, radiation and hormonal therapy is typically used to treat tumors. However, said Yolanda Brice, M.D., an interventional radiologist at Memorial Sloan Kettering Cancer Center, “when treated with only radiation and hormonal therapy, the tumors will eventually return.” Cryotherapy, Brice said, could be a more effective way to treat cancer for patients who can’t have surgery.
“The fact that we only saw a 10 percent recurrence rate in our study is incredibly promising,” she said.