Genomic Data Has a Diversity Problem, But Global Efforts Are Underway to Fix It

Genetic data sets skew too European, threatening to narrow who will benefit from future advances.
Genomics has begun its golden age. Just 20 years ago, sequencing a single genome cost nearly $3 billion and took over a decade. Today, the same feat can be achieved for a few hundred dollars and the better part of a day . Suddenly, the prospect of sequencing not just individuals, but whole populations, has become feasible.
The genetic differences between humans may seem meager, only around 0.1 percent of the genome on average, but this variation can have profound effects on an individual's risk of disease, responsiveness to medication, and even the dosage level that would work best.
Already, initiatives like the U.K.'s 100,000 Genomes Project - now expanding to 1 million genomes - and other similarly massive sequencing projects in Iceland and the U.S., have begun collecting population-scale data in order to capture and study this variation.
The resulting data sets are immensely valuable to researchers and drug developers working to design new 'precision' medicines and diagnostics, and to gain insights that may benefit patients. Yet, because the majority of this data comes from developed countries with well-established scientific and medical infrastructure, the data collected so far is heavily biased towards Western populations with largely European ancestry.
This presents a startling and fast-emerging problem: groups that are under-represented in these datasets are likely to benefit less from the new wave of therapeutics, diagnostics, and insights, simply because they were tailored for the genetic profiles of people with European ancestry.
We may indeed be approaching a golden age of genomics-enabled precision medicine. But if the data bias persists then there is a risk, as with most golden ages throughout history, that the benefits will not be equally accessible to all, and existing inequalities will only be exacerbated.
To remedy the situation, a number of initiatives have sprung up to sequence genomes of under-represented groups, adding them to the datasets and ensuring that they too will benefit from the rapidly unfolding genomic revolution.
Global Gene Corp
The idea behind Global Gene Corp was born eight years ago in Harvard when Sumit Jamuar, co-founder and CEO, met up with his two other co-founders, both experienced geneticists, for a coffee.
"They were discussing the limitless applications of understanding your genetic code," said Jamuar, a business executive from New Delhi.
"And so, being a technology enthusiast type, I was excited and I turned to them and said hey, this is incredible! Could you sequence me and give me some insights? And they actually just turned around and said no, because it's not going to be useful for you - there's not enough reference for what a good Sumit looks like."
What started as a curiosity-driven conversation on the power of genomics ended with a commitment to tackle one of the field's biggest roadblocks - its lack of global representation.
Jamuar set out to begin with India, which has about 20 percent of the world's population, including over 4000 different ethnicities, but contributes less than 2 percent of genomic data, he told Leaps.org.
Eight years later, Global Gene Corp's sequencing initiative is well underway, and is the largest in the history of the Indian subcontinent. The program is being carried out in collaboration with biotech giant Regeneron, with support from the Indian government, local communities, and the Indian healthcare ecosystem. In August 2020, Global Gene Corp's work was recognized through the $1 million 2020 Roddenberry award for organizations that advance the vision of 'Star Trek' creator Gene Roddenberry to better humanity.
This problem has already begun to manifest itself in, for example, much higher levels of genetic misdiagnosis among non-Europeans tested for their risk of certain diseases, such as hypertrophic cardiomyopathy - an inherited disease of the heart muscle.
Global Gene Corp also focuses on developing and implementing AI and machine learning tools to make sense of the deluge of genomic data. These tools are increasingly used by both industry and academia to guide future research by identifying particularly promising or clinically interesting genetic variants. But if the underlying data is skewed European, then the effectiveness of the computational analysis - along with the future advances and avenues of research that emerge from it - will be skewed towards Europeans too.
This problem has already begun to manifest itself in, for example, much higher levels of genetic misdiagnosis among non-Europeans tested for their risk of certain diseases, such as hypertrophic cardiomyopathy - an inherited disease of the heart muscle. Most of the genetic variants used in these tests were identified as being causal for the disease from studies of European genomes. However, many of these variants differ both in their distribution and clinical significance across populations, leading to many patients of non-European ancestry receiving false-positive test results - as their benign genetic variants were misclassified as pathogenic. Had even a small number of genomes from other ethnicities been included in the initial studies, these misdiagnoses could have been avoided.
"Unless we have a data set which is unbiased and representative, we're never going to achieve the success that we want," Jamuar says.
"When Siri was first launched, she could hardly recognize an accent which was not of a certain type, so if I was trying to speak to Siri, I would have to repeat myself multiple times and try to mimic an accent which wasn't my accent so that she could understand it.
"But over time the voice recognition technology improved tremendously because the training data was expanded to include people of very diverse backgrounds and their accents, so the algorithms were trained to be able to pick that up and it dramatically improved the technology. That's the way we have to think about it - without that good-quality diverse data, we will never be able to achieve the full potential of the computational tools."
While mapping India's rich genetic diversity has been the organization's primary focus so far, they plan, in time, to expand their work to other under-represented groups in Asia, the Middle East, Africa, and Latin America.
"As other like-minded people and partners join the mission, it just accelerates the achievement of what we have set out to do, which is to map out and organize the world's genomic diversity so that we can enable high-quality life and longevity benefits for everyone, everywhere," Jamuar says.
Empowering African Genomics
Africa is the birthplace of our species, and today still retains an inordinate amount of total human genetic diversity. Groups that left Africa and went on to populate the rest of the world, some 50 to 100,000 years ago, were likely small in number and only took a fraction of the total genetic diversity with them. This ancient bottleneck means that no other group in the world can match the level of genetic diversity seen in modern African populations.
Despite Africa's central importance in understanding the history and extent of human genetic diversity, the genomics of African populations remains wildly understudied. Addressing this disparity has become a central focus of the H3Africa Consortium, an initiative formally launched in 2012 with support from the African Academy of Sciences, the U.S. National Institutes of Health, and the UK's Wellcome Trust. Today, H3Africa supports over 50 projects across the continent, on an array of different research areas in genetics relevant to the health and heredity of Africans.
"Africa is the cradle of Humankind. So what that really means is that the populations that are currently living in Africa are among some of the oldest populations on the globe, and we know that the longer populations have had to go through evolutionary phases, the more variation there is in the genomes of people who live presently," says Zane Lombard, a principal investigator at H3Africa and Associate Professor of Human Genetics at the University of the Witwatersrand in Johannesburg, South Africa.
"So for that reason, African populations carry a huge amount of genetic variation and diversity, which is pretty much uncaptured. There's still a lot to learn as far as novel variation is concerned by looking at and studying African genomes."
A recent landmark H3Africa study, led by Lombard and published in Nature in October, sequenced the genomes of over 400 African individuals from 50 ethno-linguistic groups - many of which had never been sampled before.
Despite the relatively modest number of individuals sequenced in the study, over three million previously undescribed genetic variants were found, and complex patterns of ancestral migration were uncovered.
"In some of these ethno-linguistic groups they don't have a word for DNA, so we've had to really think about how to make sure that we communicate the purposes of different studies to participants so that you have true informed consent," says Lombard.
"The objective," she explained, "was to try and fill some of the gaps for many of these populations for which we didn't have any whole genome sequences or any genetic variation data...because if we're thinking about the future of precision medicine, if the patient is a member of a specific group where we don't know a lot about the genomic variation that exists in that group, it makes it really difficult to start thinking about clinical interpretation of their data."
From H3Africa's conception, the consortium's goal has not only been to better represent Africa's staggering genetic diversity in genomic data sets, but also to build Africa's domestic genomics capabilities and empower a new generation of African researchers. By doing so, the hope is that Africans will be able to set their own genomics agenda, and leapfrog to new and better ways of doing the work.
"The training that has happened on the continent and the number of new scientists, new students, and fellows that have come through the process and are now enabled to start their own research groups, to grow their own research in their countries, to be a spokesperson for genomics research in their countries, and to build that political will to do these larger types of sequencing initiatives - that is really a significant outcome from H3Africa as well. Over and above all the science that's coming out," Lombard says.
"What has been created through H3Africa is just this locus of researchers and scientists and bioethicists who have the same goal at heart - to work towards adjusting the data bias and making sure that all global populations are represented in genomics."
Dr. May Edward Chinn, Kizzmekia Corbett, PhD., and Alice Ball, among others, have been behind some of the most important scientific work of the last century.
If you look back on the last century of scientific achievements, you might notice that most of the scientists we celebrate are overwhelmingly white, while scientists of color take a backseat. Since the Nobel Prize was introduced in 1901, for example, no black scientists have landed this prestigious award.
The work of black women scientists has gone unrecognized in particular. Their work uncredited and often stolen, black women have nevertheless contributed to some of the most important advancements of the last 100 years, from the polio vaccine to GPS.
Here are five black women who have changed science forever.
Dr. May Edward Chinn
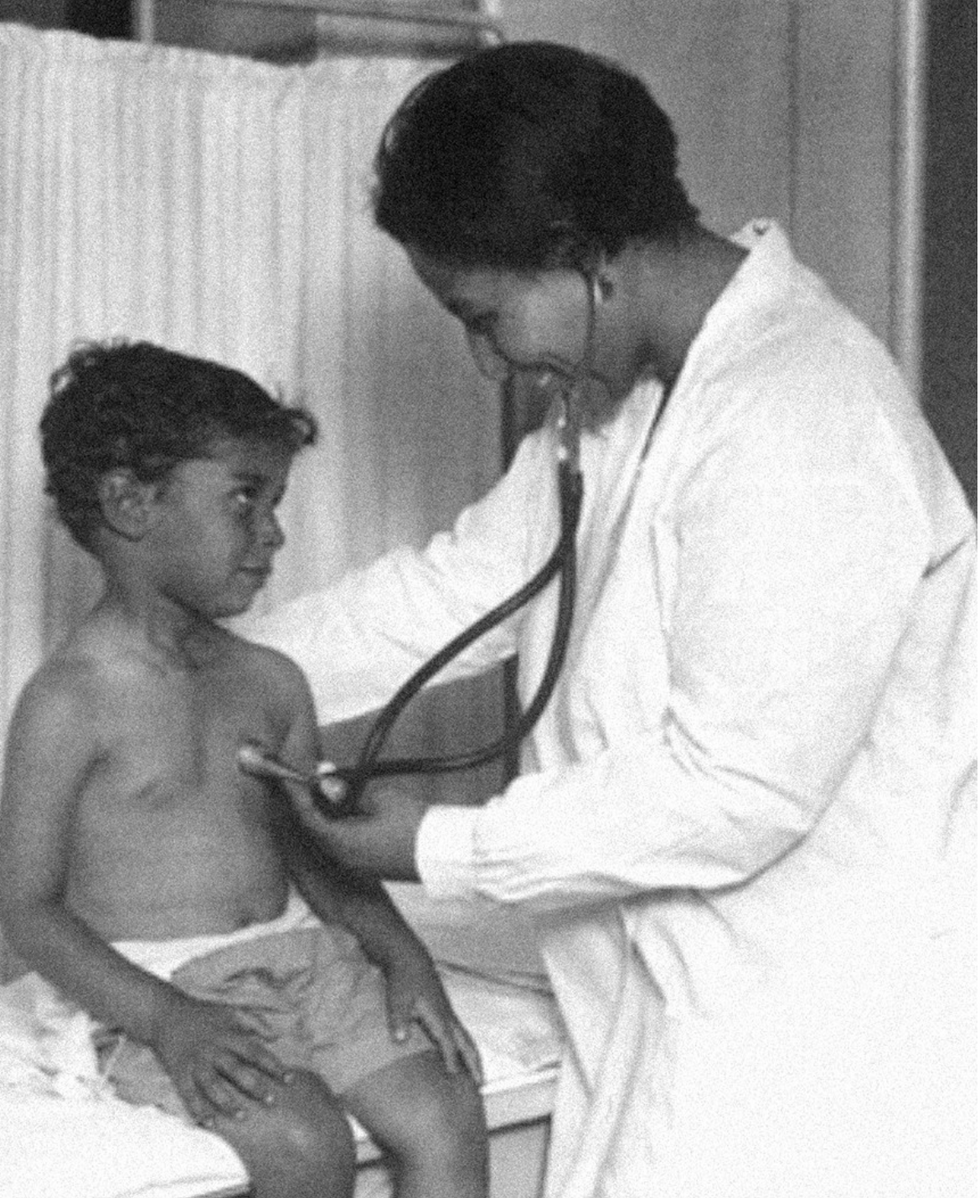
Dr. May Edward Chinn practicing medicine in Harlem
George B. Davis, PhD.
Chinn was born to poor parents in New York City just before the start of the 20th century. Although she showed great promise as a pianist, playing with the legendary musician Paul Robeson throughout the 1920s, she decided to study medicine instead. Chinn, like other black doctors of the time, were barred from studying or practicing in New York hospitals. So Chinn formed a private practice and made house calls, sometimes operating in patients’ living rooms, using an ironing board as a makeshift operating table.
Chinn worked among the city’s poor, and in doing this, started to notice her patients had late-stage cancers that often had gone undetected or untreated for years. To learn more about cancer and its prevention, Chinn begged information off white doctors who were willing to share with her, and even accompanied her patients to other clinic appointments in the city, claiming to be the family physician. Chinn took this information and integrated it into her own practice, creating guidelines for early cancer detection that were revolutionary at the time—for instance, checking patient health histories, checking family histories, performing routine pap smears, and screening patients for cancer even before they showed symptoms. For years, Chinn was the only black female doctor working in Harlem, and she continued to work closely with the poor and advocate for early cancer screenings until she retired at age 81.
Alice Ball

Pictorial Press Ltd/Alamy
Alice Ball was a chemist best known for her groundbreaking work on the development of the “Ball Method,” the first successful treatment for those suffering from leprosy during the early 20th century.
In 1916, while she was an undergraduate student at the University of Hawaii, Ball studied the effects of Chaulmoogra oil in treating leprosy. This oil was a well-established therapy in Asian countries, but it had such a foul taste and led to such unpleasant side effects that many patients refused to take it.
So Ball developed a method to isolate and extract the active compounds from Chaulmoogra oil to create an injectable medicine. This marked a significant breakthrough in leprosy treatment and became the standard of care for several decades afterward.
Unfortunately, Ball died before she could publish her results, and credit for this discovery was given to another scientist. One of her colleagues, however, was able to properly credit her in a publication in 1922.
Henrietta Lacks

onathan Newton/The Washington Post/Getty
The person who arguably contributed the most to scientific research in the last century, surprisingly, wasn’t even a scientist. Henrietta Lacks was a tobacco farmer and mother of five children who lived in Maryland during the 1940s. In 1951, Lacks visited Johns Hopkins Hospital where doctors found a cancerous tumor on her cervix. Before treating the tumor, the doctor who examined Lacks clipped two small samples of tissue from Lacks’ cervix without her knowledge or consent—something unthinkable today thanks to informed consent practices, but commonplace back then.
As Lacks underwent treatment for her cancer, her tissue samples made their way to the desk of George Otto Gey, a cancer researcher at Johns Hopkins. He noticed that unlike the other cell cultures that came into his lab, Lacks’ cells grew and multiplied instead of dying out. Lacks’ cells were “immortal,” meaning that because of a genetic defect, they were able to reproduce indefinitely as long as certain conditions were kept stable inside the lab.
Gey started shipping Lacks’ cells to other researchers across the globe, and scientists were thrilled to have an unlimited amount of sturdy human cells with which to experiment. Long after Lacks died of cervical cancer in 1951, her cells continued to multiply and scientists continued to use them to develop cancer treatments, to learn more about HIV/AIDS, to pioneer fertility treatments like in vitro fertilization, and to develop the polio vaccine. To this day, Lacks’ cells have saved an estimated 10 million lives, and her family is beginning to get the compensation and recognition that Henrietta deserved.
Dr. Gladys West

Andre West
Gladys West was a mathematician who helped invent something nearly everyone uses today. West started her career in the 1950s at the Naval Surface Warfare Center Dahlgren Division in Virginia, and took data from satellites to create a mathematical model of the Earth’s shape and gravitational field. This important work would lay the groundwork for the technology that would later become the Global Positioning System, or GPS. West’s work was not widely recognized until she was honored by the US Air Force in 2018.
Dr. Kizzmekia "Kizzy" Corbett

TIME Magazine
At just 35 years old, immunologist Kizzmekia “Kizzy” Corbett has already made history. A viral immunologist by training, Corbett studied coronaviruses at the National Institutes of Health (NIH) and researched possible vaccines for coronaviruses such as SARS (Severe Acute Respiratory Syndrome) and MERS (Middle East Respiratory Syndrome).
At the start of the COVID pandemic, Corbett and her team at the NIH partnered with pharmaceutical giant Moderna to develop an mRNA-based vaccine against the virus. Corbett’s previous work with mRNA and coronaviruses was vital in developing the vaccine, which became one of the first to be authorized for emergency use in the United States. The vaccine, along with others, is responsible for saving an estimated 14 million lives.On today’s episode of Making Sense of Science, I’m honored to be joined by Dr. Paul Song, a physician, oncologist, progressive activist and biotech chief medical officer. Through his company, NKGen Biotech, Dr. Song is leveraging the power of patients’ own immune systems by supercharging the body’s natural killer cells to make new treatments for Alzheimer’s and cancer.
Whereas other treatments for Alzheimer’s focus directly on reducing the build-up of proteins in the brain such as amyloid and tau in patients will mild cognitive impairment, NKGen is seeking to help patients that much of the rest of the medical community has written off as hopeless cases, those with late stage Alzheimer’s. And in small studies, NKGen has shown remarkable results, even improvement in the symptoms of people with these very progressed forms of Alzheimer’s, above and beyond slowing down the disease.
In the realm of cancer, Dr. Song is similarly setting his sights on another group of patients for whom treatment options are few and far between: people with solid tumors. Whereas some gradual progress has been made in treating blood cancers such as certain leukemias in past few decades, solid tumors have been even more of a challenge. But Dr. Song’s approach of using natural killer cells to treat solid tumors is promising. You may have heard of CAR-T, which uses genetic engineering to introduce cells into the body that have a particular function to help treat a disease. NKGen focuses on other means to enhance the 40 plus receptors of natural killer cells, making them more receptive and sensitive to picking out cancer cells.
Paul Y. Song, MD is currently CEO and Vice Chairman of NKGen Biotech. Dr. Song’s last clinical role was Asst. Professor at the Samuel Oschin Cancer Center at Cedars Sinai Medical Center.
Dr. Song served as the very first visiting fellow on healthcare policy in the California Department of Insurance in 2013. He is currently on the advisory board of the Pritzker School of Molecular Engineering at the University of Chicago and a board member of Mercy Corps, The Center for Health and Democracy, and Gideon’s Promise.
Dr. Song graduated with honors from the University of Chicago and received his MD from George Washington University. He completed his residency in radiation oncology at the University of Chicago where he served as Chief Resident and did a brachytherapy fellowship at the Institute Gustave Roussy in Villejuif, France. He was also awarded an ASTRO research fellowship in 1995 for his research in radiation inducible gene therapy.
With Dr. Song’s leadership, NKGen Biotech’s work on natural killer cells represents cutting-edge science leading to key findings and important pieces of the puzzle for treating two of humanity’s most intractable diseases.
Show links
- Paul Song LinkedIn
- NKGen Biotech on Twitter - @NKGenBiotech
- NKGen Website: https://nkgenbiotech.com/
- NKGen appoints Paul Song
- Patient Story: https://pix11.com/news/local-news/long-island/promising-new-treatment-for-advanced-alzheimers-patients/
- FDA Clearance: https://nkgenbiotech.com/nkgen-biotech-receives-ind-clearance-from-fda-for-snk02-allogeneic-natural-killer-cell-therapy-for-solid-tumors/Q3 earnings data: https://www.nasdaq.com/press-release/nkgen-biotech-inc.-reports-third-quarter-2023-financial-results-and-business