Blood Test Can Detect Lymphoma Cells Before a Tumor Grows Back
David Kurtz making DNA sequencing libraries in his lab.
When David M. Kurtz was doing his clinical fellowship at Stanford University Medical Center in 2009, specializing in lymphoma treatments, he found himself grappling with a question no one could answer. A typical regimen for these blood cancers prescribed six cycles of chemotherapy, but no one knew why. "The number seemed to be drawn out of a hat," Kurtz says. Some patients felt much better after just two doses, but had to endure the toxic effects of the entire course. For some elderly patients, the side effects of chemo are so harsh, they alone can kill. Others appeared to be cancer-free on the CT scans after the requisite six but then succumbed to it months later.
"Anecdotally, one patient decided to stop therapy after one dose because he felt it was so toxic that he opted for hospice instead," says Kurtz, now an oncologist at the center. "Five years down the road, he was alive and well. For him, just one dose was enough." Others would return for their one-year check up and find that their tumors grew back. Kurtz felt that while CT scans and MRIs were powerful tools, they weren't perfect ones. They couldn't tell him if there were any cancer cells left, stealthily waiting to germinate again. The scans only showed the tumor once it was back.
Blood cancers claim about 68,000 people a year, with a new diagnosis made about every three minutes, according to the Leukemia Research Foundation. For patients with B-cell lymphoma, which Kurtz focuses on, the survival chances are better than for some others. About 60 percent are cured, but the remaining 40 percent will relapse—possibly because they will have a negative CT scan, but still harbor malignant cells. "You can't see this on imaging," says Michael Green, who also treats blood cancers at University of Texas MD Anderson Medical Center.
The new blood test is sensitive enough to spot one cancerous perpetrator amongst one million other DNA molecules.
Kurtz wanted a better diagnostic tool, so he started working on a blood test that could capture the circulating tumor DNA or ctDNA. For that, he needed to identify the specific mutations typical for B-cell lymphomas. Working together with another fellow PhD student Jake Chabon, Kurtz finally zeroed-in on the tumor's genetic "appearance" in 2017—a pair of specific mutations sitting in close proximity to each other—a rare and telling sign. The human genome contains about 3 billion base pairs of nucleotides—molecules that compose genes—and in case of the B-cell lymphoma cells these two mutations were only a few base pairs apart. "That was the moment when the light bulb went on," Kurtz says.
The duo formed a company named Foresight Diagnostics, focusing on taking the blood test to the clinic. But knowing the tumor's mutational signature was only half the process. The other was fishing the tumor's DNA out of patients' bloodstream that contains millions of other DNA molecules, explains Chabon, now Foresight's CEO. It would be like looking for an escaped criminal in a large crowd. Kurtz and Chabon solved the problem by taking the tumor's "mug shot" first. Doctors would take the biopsy pre-treatment and sequence the tumor, as if taking the criminal's photo. After treatments, they would match the "mug shot" to all DNA molecules derived from the patient's blood sample to see if any molecular criminals managed to escape the chemo.
Foresight isn't the only company working on blood-based tumor detection tests, which are dubbed liquid biopsies—other companies such as Natera or ArcherDx developed their own. But in a recent study, the Foresight team showed that their method is significantly more sensitive in "fishing out" the cancer molecules than existing tests. Chabon says that this test can detect circulating tumor DNA in concentrations that are nearly 100 times lower than other methods. Put another way, it's sensitive enough to spot one cancerous perpetrator amongst one million other DNA molecules.
They also aim to extend their test to detect other malignancies such as lung, breast or colorectal cancers.
"It increases the sensitivity of detection and really catches most patients who are going to progress," says Green, the University of Texas oncologist who wasn't involved in the study, but is familiar with the method. It would also allow monitoring patients during treatment and making better-informed decisions about which therapy regimens would be most effective. "It's a minimally invasive test," Green says, and "it gives you a very high confidence about what's going on."
Having shown that the test works well, Kurtz and Chabon are planning a new trial in which oncologists would rely on their method to decide when to stop or continue chemo. They also aim to extend their test to detect other malignancies such as lung, breast or colorectal cancers. The latest genome sequencing technologies have sequenced and catalogued over 2,500 different tumor specimens and the Foresight team is analyzing this data, says Chabon, which gives the team the opportunity to create more molecular "mug shots."
The team hopes that that their blood cancer test will become available to patients within about five years, making doctors' job easier, and not only at the biological level. "When I tell patients, "good news, your cancer is in remission', they ask me, 'does it mean I'm cured?'" Kurtz says. "Right now I can't answer this question because I don't know—but I would like to." His company's test, he hopes, will enable him to reply with certainty. He'd very much like to have the power of that foresight.
This article is republished from our archives to coincide with Blood Cancer Awareness Month, which highlights progress in cancer diagnostics and treatment.
Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.
Researchers probe extreme gene therapy for severe alcoholism
When all traditional therapeutic approaches fail for alcohol abuse disorder, a radical gene therapy might be something to try in the future.
Story by Freethink
A single shot — a gene therapy injected into the brain — dramatically reduced alcohol consumption in monkeys that previously drank heavily. If the therapy is safe and effective in people, it might one day be a permanent treatment for alcoholism for people with no other options.
The challenge: Alcohol use disorder (AUD) means a person has trouble controlling their alcohol consumption, even when it is negatively affecting their life, job, or health.
In the U.S., more than 10 percent of people over the age of 12 are estimated to have AUD, and while medications, counseling, or sheer willpower can help some stop drinking, staying sober can be a huge struggle — an estimated 40-60 percent of people relapse at least once.
A team of U.S. researchers suspected that an in-development gene therapy for Parkinson’s disease might work as a dopamine-replenishing treatment for alcoholism, too.
According to the CDC, more than 140,000 Americans are dying each year from alcohol-related causes, and the rate of deaths has been rising for years, especially during the pandemic.
The idea: For occasional drinkers, alcohol causes the brain to release more dopamine, a chemical that makes you feel good. Chronic alcohol use, however, causes the brain to produce, and process, less dopamine, and this persistent dopamine deficit has been linked to alcohol relapse.
There is currently no way to reverse the changes in the brain brought about by AUD, but a team of U.S. researchers suspected that an in-development gene therapy for Parkinson’s disease might work as a dopamine-replenishing treatment for alcoholism, too.
To find out, they tested it in heavy-drinking monkeys — and the animals’ alcohol consumption dropped by 90% over the course of a year.
How it works: The treatment centers on the protein GDNF (“glial cell line-derived neurotrophic factor”), which supports the survival of certain neurons, including ones linked to dopamine.
For the new study, a harmless virus was used to deliver the gene that codes for GDNF into the brains of four monkeys that, when they had the option, drank heavily — the amount of ethanol-infused water they consumed would be equivalent to a person having nine drinks per day.
“We targeted the cell bodies that produce dopamine with this gene to increase dopamine synthesis, thereby replenishing or restoring what chronic drinking has taken away,” said co-lead researcher Kathleen Grant.
To serve as controls, another four heavy-drinking monkeys underwent the same procedure, but with a saline solution delivered instead of the gene therapy.
The results: All of the monkeys had their access to alcohol removed for two months following the surgery. When it was then reintroduced for four weeks, the heavy drinkers consumed 50 percent less compared to the control group.
When the researchers examined the monkeys’ brains at the end of the study, they were able to confirm that dopamine levels had been replenished in the treated animals, but remained low in the controls.
The researchers then took the alcohol away for another four weeks, before giving it back for four. They repeated this cycle for a year, and by the end of it, the treated monkeys’ consumption had fallen by more than 90 percent compared to the controls.
“Drinking went down to almost zero,” said Grant. “For months on end, these animals would choose to drink water and just avoid drinking alcohol altogether. They decreased their drinking to the point that it was so low we didn’t record a blood-alcohol level.”
When the researchers examined the monkeys’ brains at the end of the study, they were able to confirm that dopamine levels had been replenished in the treated animals, but remained low in the controls.
Looking ahead: Dopamine is involved in a lot more than addiction, so more research is needed to not only see if the results translate to people but whether the gene therapy leads to any unwanted changes to mood or behavior.
Because the therapy requires invasive brain surgery and is likely irreversible, it’s unlikely to ever become a common treatment for alcoholism — but it could one day be the only thing standing between people with severe AUD and death.
“[The treatment] would be most appropriate for people who have already shown that all our normal therapeutic approaches do not work for them,” said Grant. “They are likely to create severe harm or kill themselves or others due to their drinking.”
This article originally appeared on Freethink, home of the brightest minds and biggest ideas of all time.

DNA- and RNA-based electronic implants may revolutionize healthcare
The test tubes contain tiny DNA/enzyme-based circuits, which comprise TRUMPET, a new type of electronic device, smaller than a cell.
Implantable electronic devices can significantly improve patients’ quality of life. A pacemaker can encourage the heart to beat more regularly. A neural implant, usually placed at the back of the skull, can help brain function and encourage higher neural activity. Current research on neural implants finds them helpful to patients with Parkinson’s disease, vision loss, hearing loss, and other nerve damage problems. Several of these implants, such as Elon Musk’s Neuralink, have already been approved by the FDA for human use.
Yet, pacemakers, neural implants, and other such electronic devices are not without problems. They require constant electricity, limited through batteries that need replacements. They also cause scarring. “The problem with doing this with electronics is that scar tissue forms,” explains Kate Adamala, an assistant professor of cell biology at the University of Minnesota Twin Cities. “Anytime you have something hard interacting with something soft [like muscle, skin, or tissue], the soft thing will scar. That's why there are no long-term neural implants right now.” To overcome these challenges, scientists are turning to biocomputing processes that use organic materials like DNA and RNA. Other promised benefits include “diagnostics and possibly therapeutic action, operating as nanorobots in living organisms,” writes Evgeny Katz, a professor of bioelectronics at Clarkson University, in his book DNA- And RNA-Based Computing Systems.
While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output.
Adamala’s research focuses on developing such biocomputing systems using DNA, RNA, proteins, and lipids. Using these molecules in the biocomputing systems allows the latter to be biocompatible with the human body, resulting in a natural healing process. In a recent Nature Communications study, Adamala and her team created a new biocomputing platform called TRUMPET (Transcriptional RNA Universal Multi-Purpose GatE PlaTform) which acts like a DNA-powered computer chip. “These biological systems can heal if you design them correctly,” adds Adamala. “So you can imagine a computer that will eventually heal itself.”
The basics of biocomputing
Biocomputing and regular computing have many similarities. Like regular computing, biocomputing works by running information through a series of gates, usually logic gates. A logic gate works as a fork in the road for an electronic circuit. The input will travel one way or another, giving two different outputs. An example logic gate is the AND gate, which has two inputs (A and B) and two different results. If both A and B are 1, the AND gate output will be 1. If only A is 1 and B is 0, the output will be 0 and vice versa. If both A and B are 0, the result will be 0. While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output. In this case, the DNA enters the logic gate as a single or double strand.
If the DNA is double-stranded, the system “digests” the DNA or destroys it, which results in non-fluorescence or “0” output. Conversely, if the DNA is single-stranded, it won’t be digested and instead will be copied by several enzymes in the biocomputing system, resulting in fluorescent RNA or a “1” output. And the output for this type of binary system can be expanded beyond fluorescence or not. For example, a “1” output might be the production of the enzyme insulin, while a “0” may be that no insulin is produced. “This kind of synergy between biology and computation is the essence of biocomputing,” says Stephanie Forrest, a professor and the director of the Biodesign Center for Biocomputing, Security and Society at Arizona State University.
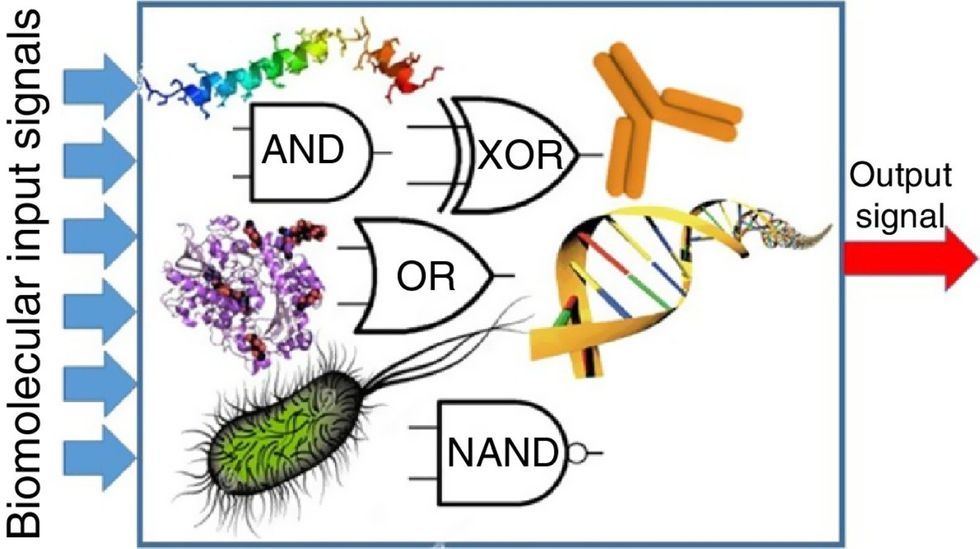
Biocomputing circles are made of DNA, RNA, proteins and even bacteria.
Evgeny Katz
The TRUMPET’s promise
Depending on whether the biocomputing system is placed directly inside a cell within the human body, or run in a test-tube, different environmental factors play a role. When an output is produced inside a cell, the cell's natural processes can amplify this output (for example, a specific protein or DNA strand), creating a solid signal. However, these cells can also be very leaky. “You want the cells to do the thing you ask them to do before they finish whatever their businesses, which is to grow, replicate, metabolize,” Adamala explains. “However, often the gate may be triggered without the right inputs, creating a false positive signal. So that's why natural logic gates are often leaky." While biocomputing outside a cell in a test tube can allow for tighter control over the logic gates, the outputs or signals cannot be amplified by a cell and are less potent.
TRUMPET, which is smaller than a cell, taps into both cellular and non-cellular biocomputing benefits. “At its core, it is a nonliving logic gate system,” Adamala states, “It's a DNA-based logic gate system. But because we use enzymes, and the readout is enzymatic [where an enzyme replicates the fluorescent RNA], we end up with signal amplification." This readout means that the output from the TRUMPET system, a fluorescent RNA strand, can be replicated by nearby enzymes in the platform, making the light signal stronger. "So it combines the best of both worlds,” Adamala adds.
These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body.
The TRUMPET biocomputing process is relatively straightforward. “If the DNA [input] shows up as single-stranded, it will not be digested [by the logic gate], and you get this nice fluorescent output as the RNA is made from the single-stranded DNA, and that's a 1,” Adamala explains. "And if the DNA input is double-stranded, it gets digested by the enzymes in the logic gate, and there is no RNA created from the DNA, so there is no fluorescence, and the output is 0." On the story's leading image above, if the tube is "lit" with a purple color, that is a binary 1 signal for computing. If it's "off" it is a 0.
While still in research, TRUMPET and other biocomputing systems promise significant benefits to personalized healthcare and medicine. These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body. The study’s lead author and graduate student Judee Sharon is already beginning to research TRUMPET's ability for earlier cancer diagnoses. Because the inputs for TRUMPET are single or double-stranded DNA, any mutated or cancerous DNA could theoretically be detected from the platform through the biocomputing process. Theoretically, devices like TRUMPET could be used to detect cancer and other diseases earlier.
Adamala sees TRUMPET not only as a detection system but also as a potential cancer drug delivery system. “Ideally, you would like the drug only to turn on when it senses the presence of a cancer cell. And that's how we use the logic gates, which work in response to inputs like cancerous DNA. Then the output can be the production of a small molecule or the release of a small molecule that can then go and kill what needs killing, in this case, a cancer cell. So we would like to develop applications that use this technology to control the logic gate response of a drug’s delivery to a cell.”
Although platforms like TRUMPET are making progress, a lot more work must be done before they can be used commercially. “The process of translating mechanisms and architecture from biology to computing and vice versa is still an art rather than a science,” says Forrest. “It requires deep computer science and biology knowledge,” she adds. “Some people have compared interdisciplinary science to fusion restaurants—not all combinations are successful, but when they are, the results are remarkable.”