A Cancer Researcher Opens Up About His Astonishing Breakthrough
Kira Peikoff was the editor-in-chief of Leaps.org from 2017 to 2021. As a journalist, her work has appeared in The New York Times, Newsweek, Nautilus, Popular Mechanics, The New York Academy of Sciences, and other outlets. She is also the author of four suspense novels that explore controversial issues arising from scientific innovation: Living Proof, No Time to Die, Die Again Tomorrow, and Mother Knows Best. Peikoff holds a B.A. in Journalism from New York University and an M.S. in Bioethics from Columbia University. She lives in New Jersey with her husband and two young sons. Follow her on Twitter @KiraPeikoff.
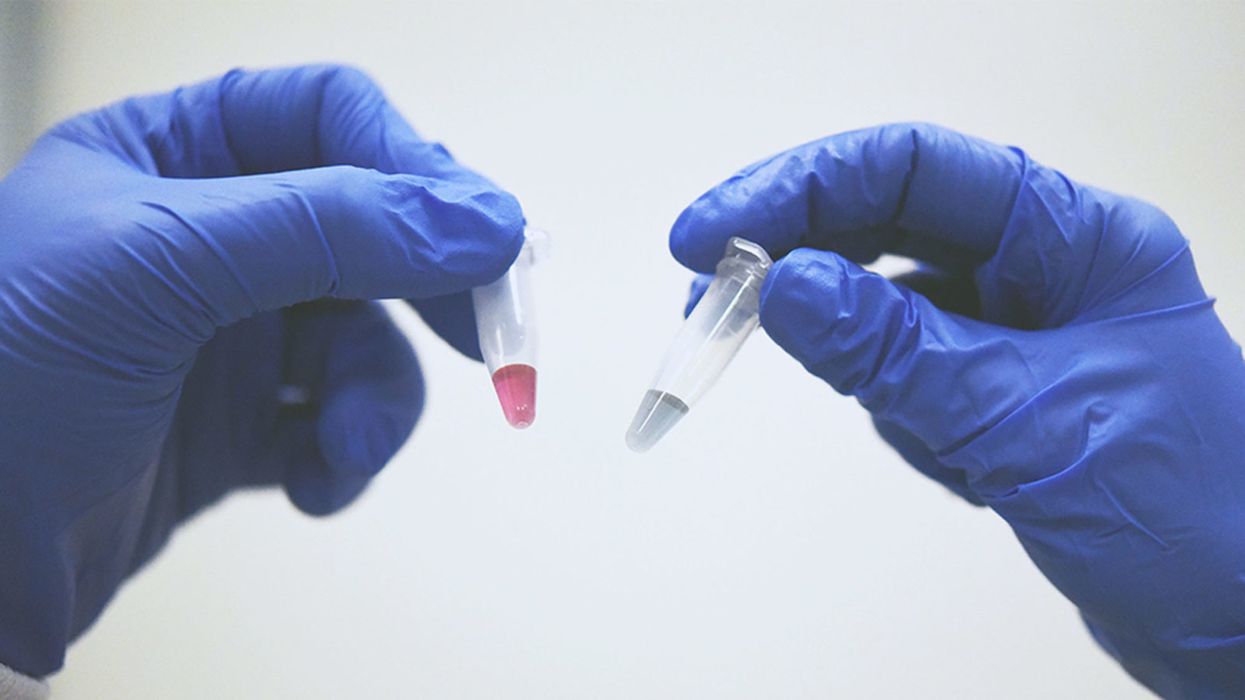
A simple ten-minute universal cancer test that can be detected by the human eye or an electronic device - published in Nature Communications (Dec 2018) by the Trau lab at the University of Queensland. Red indicates the presence of cancerous cells and blue doesn't.
Matt Trau, a professor of chemistry at the University of Queensland, stunned the science world back in December when the prestigious journal Nature Communications published his lab's discovery about a unique property of cancer DNA that could lead to a simple, cheap, and accurate test to detect any type of cancer in under 10 minutes.
No one believed it. I didn't believe it. I thought, "Gosh, okay, maybe it's a fluke."
Trau granted very few interviews in the wake of the news, but he recently opened up to leapsmag about the significance of this promising early research. Here is his story in his own words, as told to Editor-in-Chief Kira Peikoff.
There's been an incredible explosion of knowledge over the past 20 years, particularly since the genome was sequenced. The area of diagnostics has a tremendous amount of promise and has caught our lab's interest. If you catch cancer early, you can improve survival rates to as high as 98 percent, sometimes even now surpassing that.
My lab is interested in devices to improve the trajectory of cancer patients. So, once people get diagnosed, can we get really sophisticated information about the molecular origins of the disease, and can we measure it in real time? And then can we match that with the best treatment and monitor it in real time, too?
I think those approaches, also coupled with immunotherapy, where one dreams of monitoring the immune system simultaneously with the disease progress, will be the future.
But currently, the methodologies for cancer are still pretty old. So, for example, let's talk about biopsies in general. Liquid biopsy just means using a blood test or a urine test, rather than extracting out a piece of solid tissue. Now consider breast cancer. Still, the cutting-edge screening method is mammography or the physical interrogation for lumps. This has had a big impact in terms of early detection and awareness, but it's still primitive compared to interrogating, forensically, blood samples to look at traces of DNA.
Large machines like CAT scans, PET scans, MRIs, are very expensive and very subjective in terms of the operator. They don't look at the root causes of the cancer. Cancer is caused by changes in DNA. These can be changes in the hard drive of the DNA (the genomic changes) or changes in the apps that the DNA are running (the epigenetics and the transcriptomics).
We don't look at that now, even though we have, emerging, all of these technologies to do it, and those technologies are getting so much cheaper. I saw some statistics at a conference just a few months ago that, in the United States, less than 1 percent of cancer patients have their DNA interrogated. That's the current state-of-the-art in the modern medical system.

Professor Matt Trau, a cancer researcher at the University of Queensland in Australia.
(Courtesy)
Blood, as the highway of the body, is carrying all of this information. Cancer cells, if they are present in the body, are constantly getting turned over. When they die, they release their contents into the blood. Many of these cells end up in the urine and saliva. Having technologies that can forensically scan the highways looking for evidence of cancer is little bit like looking for explosives at the airport. That's very valuable as a security tool.
The trouble is that there are thousands of different types of cancer. Going back to breast cancer, there's at least a dozen different types, probably more, and each of them change the DNA (the hard drive of the disease) and the epigenetics (or the RAM memory). So one of the problems for diagnostics in cancer is to find something that is a signature of all cancers. That's been a really, really, really difficult problem.
Ours was a completely serendipitous discovery. What we found in the lab was this one marker that just kept coming up in all of the types of breast cancers we were studying.
No one believed it. I didn't believe it. I thought, "Gosh, okay, maybe it's a fluke, maybe it works just for breast cancer." So we went on to test it in prostate cancer, which is also many different types of diseases, and it seemed to be working in all of those. We then tested it further in lymphoma. Again, many different types of lymphoma. It worked across all of those. We tested it in gastrointestinal cancer. Again, many different types, and still, it worked, but we were skeptical.
Then we looked at cell lines, which are cells that have come from previous cancer patients, that we grow in the lab, but are used as model experimental systems. We have many of those cell lines, both ones that are cancerous, and ones that are healthy. It was quite remarkable that the marker worked in all of the cancer cell lines and didn't work in the healthy cell lines.
What could possibly be going on?
Well, imagine DNA as a piece of string, that's your hard drive. Epigenetics is like the beads that you put on that string. Those beads you can take on and off as you wish and they control which apps are run, meaning which genetic programs the cell runs. We hypothesized that for cancer, those beads cluster together, rather than being randomly distributed across the string.
Ultimately, I see this as something that would be like a pregnancy test you could take at your doctor's office.
The implications of this are profound. It means that DNA from cancer folds in water into three-dimensional structures that are very different from healthy cells' DNA. It's quite literally the needle in a haystack. Because when you do a liquid biopsy for early detection of cancer, most of the DNA from blood contains a vast abundance of healthy DNA. And that's not of interest. What's of interest is to find the cancerous DNA. That's there only in trace.
Once we figured out what was going on, we could easily set up a system to detect the trace cancerous DNA. It binds to gold nanoparticles in water and changes color. The test takes 10 minutes, and you can detect it by eye. Red indicates cancer and blue doesn't.
We're very, very excited about where we go from here. We're starting to test the test on a greater number of cancers, in thousands of patient samples. We're looking to the scientific community to engage with us, and we're getting a really good response from groups around the world who are supplying more samples to us so we can test this more broadly.
We also are very interested in testing how early can we go with this test. Can we detect cancer through a simple blood test even before there are any symptoms whatsoever? If so, we might be able to convert a cancer diagnosis to something almost as good as a vaccine.
Of course, we have to watch what are called false positives. We don't want to be detecting people as positives when they don't have cancer, and so the technology needs to improve there. We see this version as the iPhone 1. We're interested in the iPhone 2, 3, 4, getting better and better.
Ultimately, I see this as something that would be like a pregnancy test you could take at your doctor's office. If it came back positive, your doctor could say, "Look, there's some news here, but actually, it's not bad news, it's good news. We've caught this so early that we will be able to manage this, and this won't be a problem for you."
If this were to be in routine use in the medical system, countless lives could be saved. Cancer is now becoming one of the biggest killers in the world. We're talking millions upon millions upon millions of people who are affected. This really motivates our work. We might make a difference there.
Kira Peikoff was the editor-in-chief of Leaps.org from 2017 to 2021. As a journalist, her work has appeared in The New York Times, Newsweek, Nautilus, Popular Mechanics, The New York Academy of Sciences, and other outlets. She is also the author of four suspense novels that explore controversial issues arising from scientific innovation: Living Proof, No Time to Die, Die Again Tomorrow, and Mother Knows Best. Peikoff holds a B.A. in Journalism from New York University and an M.S. in Bioethics from Columbia University. She lives in New Jersey with her husband and two young sons. Follow her on Twitter @KiraPeikoff.
Based on recent research, new therapies could promote a mix of viruses in the intestines to help prevent diseases of aging.
Story by Big Think
Our gut microbiome plays a substantial role in our health and well-being. Most research, however, focuses on bacteria, rather than the viruses that hide within them. Now, research from the University of Copenhagen, newly published in Nature Microbiology, found that people who live past age 100 have a greater diversity of bacteria-infecting viruses in their intestines than younger people. Furthermore, they found that the viruses are linked to changes in bacterial metabolism that may support mucosal integrity and resistance to pathogens.
The microbiota and aging
In the early 1970s, scientists discovered that the composition of our gut microbiota changes as we age. Recent studies have found that the changes are remarkably predictable and follow a pattern: The microbiota undergoes rapid, dramatic changes as toddlers transition to solid foods; further changes become less dramatic during childhood as the microbiota strikes a balance between the host and the environment; and as that balance is achieved, the microbiota remains mostly stable during our adult years (ages 18-60). However, that stability is lost as we enter our elderly years, and the microbiome undergoes dramatic reorganization. This discovery led scientists to question what causes this change and what effect it has on health.
Centenarians have a distinct gut community enriched in microorganisms that synthesize potent antimicrobial molecules that can kill multidrug-resistant pathogens.
“We are always eager to find out why some people live extremely long lives. Previous research has shown that the intestinal bacteria of old Japanese citizens produce brand-new molecules that make them resistant to pathogenic — that is, disease-promoting — microorganisms. And if their intestines are better protected against infection, well, then that is probably one of the things that cause them to live longer than others,” said Joachim Johansen, a researcher at the University of Copenhagen.
In 2021, a team of Japanese scientists set out to characterize the effect of this change on older people’s health. They specifically wanted to determine if people who lived to be over 100 years old — that is, centenarians — underwent changes that provided them with unique benefits. They discovered centenarians have a distinct gut community enriched in microorganisms that synthesize potent antimicrobial molecules that can kill multidrug-resistant pathogens, including Clostridioides difficile and Enterococcus faecium. In other words, the late-life shift in microbiota reduces an older person’s susceptibility to common gut pathogens.
Viruses can change alter the genes of bacteria
Although the late-in-life microbiota change could be beneficial to health, it remained unclear what facilitated this shift. To solve this mystery, Johansen and his colleagues turned their attention to an often overlooked member of the microbiome: viruses. “Our intestines contain billions of viruses living inside bacteria, and they could not care less about human cells; instead, they infect the bacterial cells. And seeing as there are hundreds of different types of bacteria in our intestines, there are also lots of bacterial viruses,” said Simon Rasmussen, Johansen’s research advisor.
Centenarians had a more diverse virome, including previously undescribed viral genera.
For decades, scientists have explored the possibility of phage therapy — that is, using viruses that infect bacteria (called bacteriophages or simply phages) to kill pathogens. However, bacteriophages can also enhance the bacteria they infect. For example, they can provide genes that help their bacterial host attack other bacteria or provide new metabolic capabilities. Both of these can change which bacteria colonize the gut and, in turn, protect against certain disease states.
Intestinal viruses give bacteria new abilities
Johansen and his colleagues were interested in what types of viruses centenarians had in their gut and whether those viruses carried genes that altered metabolism. They compared fecal samples of healthy centenarians (100+ year-olds) with samples from younger patients (18-100 year-olds). They found that the centenarians had a more diverse virome, including previously undescribed viral genera.
They also revealed an enrichment of genes supporting key steps in the sulfate metabolic pathway. The authors speculate that this translates to increased levels of microbially derived sulfide, which may lead to health-promoting outcomes, such as supporting mucosal integrity and resistance to potential pathogens.
“We have learned that if a virus pays a bacterium a visit, it may actually strengthen the bacterium. The viruses we found in the healthy Japanese centenarians contained extra genes that could boost the bacteria,” said Johansen.
Simon Rasmussen added, “If you discover bacteria and viruses that have a positive effect on the human intestinal flora, the obvious next step is to find out whether only some or all of us have them. If we are able to get these bacteria and their viruses to move in with the people who do not have them, more people could benefit from them.”
This article originally appeared on Big Think, home of the brightest minds and biggest ideas of all time.
Sign up for Big Think’s newsletter

Embrace the mess: how to choose which scientists to trust
A dozen bioethicists and researchers shared their advice on how to spot the scientists searching for the truth more than money, ego or fame.
It’s no easy task these days for people to pick the scientists they should follow. According to a recent poll by NORC at the University of Chicago, only 39 percent of Americans have a "great deal" of confidence in the scientific community. The finding is similar to Pew research last year showing that 29 percent of Americans have this level of confidence in medical scientists.
Not helping: All the money in science. Just 20 percent of Pew’s survey respondents think scientists are transparent about conflicts of interest with industry. While this issue is common to many fields, the recent gold rush to foot the bill for research on therapies for healthy aging may be contributing to the overall sense of distrust. “There’s a feeling that at some point, the FDA may actually designate aging as a disease,” said Pam Maher, a neuroscientist who studies aging at Salk Institute. “That may be another impetus for a lot of these companies to start up.”
But partnering with companies is an important incentive for researchers across biomedical fields. Many scientists – with and without financial ties and incentives – are honest, transparent and doing important, inspiring work. I asked more than a dozen bioethicists and researchers in aging how to spot the scientists who are searching for the truth more than money, ego or fame.
Avoid Scientists Who Sound Overly Confident in messaging to the public. Some multi-talented scientists are adept at publishing in both top journals and media outlets. They’re great at dropping science without the confusing jargon, in ways the public can enjoy and learn from.
But do they talk in simple soundbites, painting scientific debates in pastels or black and white when colleagues use shades of gray? Maybe they crave your attention more than knowledge seeking. “When scientists speak in a very unnuanced way, that can be irresponsible,” said Josephine Johnston, a bioethicist at the Hastings Center.
Scientists should avoid exaggerations like “without a doubt” and even “we know” – unless they absolutely do. “I feel like there’s more and more hyperbole and attention seeking…[In aging research,] the loudest voices in the room are the fringe people,” said the biogenerontologist Matt Kaeberlein.
Separate Hype from Passion. Scientists should be, need to be passionate, Johnston explained. In the realm of aging, for example, Leonard Guarente, an MIT biologist and pioneer in the field of aging, told me about his belief that longer lifespans would make for a better world.
Instead of expecting scientists to be lab-dwelling robots, we should welcome their passion. It fuels scientific dedication and creativity. Fields like aging, AI and gene editing inspire the imaginations of the public and scientists alike. That’s not a bad thing.
But it does lay fertile ground for overstatements, such as claims by some that the first 1,000-year-old has already been born. If it sounds like sci-fi, it’s probably sci-fi.
Watch Out for Cult Behavior, some experts told me. Follow scientists who mix it up and engage in debates, said NYU bioethicist Arthur Caplan, not those who hang out only with researchers in the same ideological camp.
Look for whether they’re open to working with colleagues who don’t share their views. Through collaboration, they can resolve conflicting study results and data, said Danica Chen, a biologist at UC Berkeley. We should trust science as long as it doesn’t trust itself.
Messiness is Good. You want to find and follow scientists who’ve published research over the years that does not tell a clean story. “Our goal is to disprove our models,” Kaeberlein said. Scientific findings and views should zig and zag as their careers – and science – progress.
Follow scientists who write and talk publicly about new evidence that’s convinced them to reevaluate their own positions. Who embrace the inherent messiness of science – that’s the hallmark of an honest researcher.
The flipside is a very linear publishing history. Some scientists have a pet theory they’ve managed to support with more and more evidence over time, like a bricklayer gradually, flawlessly building the prettiest house in the neighborhood. Too pretty.
There’s a dark side to this charming simplicity: scientists sometimes try and succeed at engineering the very findings they’re hoping to get, said Charles Brenner, a biochemist at City of Hope National Medical Center.
These scientists “try to prove their model and ignore data that doesn’t fit their model because everybody likes a clean story,” Kaeberlein said. “People want to become famous,” said Samuel Klein, a biologist at Washington University. “So there’s always that bias to try to get positive results.”
Don’t Overvalue Credentials. Just because a scientist works at a top university doesn’t mean they’re completely trustworthy. “The institution means almost nothing,” Kaeberlein said.
Same goes for publishing in top journals, Kaeberlein added. “There’s an incentive structure that favors poor quality science and irreproducible results in high profile journals.”
Traditional proxies for credibility aren’t quite as reliable these days. Shortcuts don’t cut it anymore; you’ve got to scrutinize the actual research the scientist is producing. “You have to look at the literature and try to interpret it for yourself,” said Rafael de Cabo, a scientist at the National Institute on Aging, run by the U.S. National Institutes of Health. Or find journalists you trust to distill this information for you, Klein suggested.
Consider Company Ties. Companies can help scientists bring their research to the public more directly and efficiently than the slower grind of academia, where “the opportunities and challenges weren’t big enough for me,” said Kaeberlein, who left the University of Washington earlier this year.
"It’s generally not universities that can take technology through what we call the valley of death,” Brenner said. “There are rewards associated with taking risks.”
Many scientists are upfront about their financial conflicts of interest – sometimes out of necessity. “At a place like Duke, our conflicts of interest are very closely managed, said Matthew Hirschey, who researchers metabolism at Duke’s Molecular Physiology Institute. “We have to be incredibly explicit about our partnerships.”
But the willingness to disclose conflicts doesn’t necessarily mean the scientist is any less biased. Those conflicts can still affect their views and outcomes of their research, said Johnston, the Hastings bioethicist.
“The proof is in the pudding, and it’s got to be done by people who are not vested in making money off the results,” Klein said. Worth noting: even if scientists eschew companies, they’re almost always financially motivated to get grants for their research.
Bottom line: lots of scientists work for and with companies, and many are highly trustworthy leaders in their fields. But if a scientist is in thick with companies and checks some of the other boxes on this list, their views and research may be compromised.

