The Brave New World of Using DNA to Store Data

A panoramic view of DNA
Netscape co-founder-turned-venture capitalist billionaire investor Marc Andreessen once posited that software was eating the world. He was right, and the takeover of software resulted in many things. One of them is data. Lots and lots and lots of data. In the previous two years, humanity created more data than it did during its entire existence combined, and the amount will only increase. Think about it: The hundreds of 50KB emails you write a day, the dozens of 10MB photos, the minute-long, 350MB 4K video you shoot on your iPhone X add up to vast quantities of information. All that information needs to be stored. And that's becoming an issue as data volume outpaces storage space.
The race is on to find another medium capable of storing massive amounts of information in as small a space as possible.
"There won't be enough silicon to store all the data we need. It's unlikely that we can make flash memory smaller. We have reached the physical limits," Victor Zhirnov, chief scientist at the Semiconductor Research Corporation, says. "We are facing a crisis that's comparable to the oil crisis in the 1970s. By 2050, we're going to need to store 10 to the 30 bits, compared to 10 to the 23 bits in 2016." That amount of storage space is equivalent to each of the world's seven billion people owning almost six trillion -- that's 10 to the 12th power -- iPhone Xs with 256GB storage space.
The race is on to find another medium capable of storing massive amounts of information in as small a space as possible. Zhirnov and other scientists are looking at the human body, looking to DNA. "Nature has nailed it," Luis Ceze, a professor in the Department of Computer Science and Engineering at the University of Washington, says. "DNA is a molecular storage medium that is remarkable. It's incredibly dense, many, many thousands of times denser than the densest technology that we have today. And DNA is remarkably general. Any information you can map in bits you can store in DNA." It's so dense -- able to store a theoretical maximum of 215 petabytes (215 million gigabytes) in a single gram -- that all the data ever produced could be stored in the back of a tractor trailer truck.
Writing DNA can be an energy-efficient process, too. Consider how the human body is constantly writing and rewriting DNA, and does so on a couple thousand calories a day. And all it needs for storage is a cool, dark place, a significant energy savings when compared to server farms that require huge amounts of energy to run and even more energy to cool.
Picture it: tiny specks of inert DNA made from silicon or another material, stored in cool, dark, dry areas, preserved for all time.
Researchers first succeeded in encoding data onto DNA in 2012, when Harvard University geneticists George Church and Sri Kosuri wrote a 52,000-word book on A, C, G, and T base pairs. Their method only produced 1.28 petabytes per gram of DNA, however, a volume exceeded the next year when a group encoded all 154 Shakespeare sonnets and a 26-second clip of Martin Luther King's "I Have A Dream" speech. In 2017, Columbia University researchers Yaniv Erlich and Dina Zielinski made the process 60 percent more efficient.
The limiting factor today is cost. Erlich said the work his team did cost $7,000 to encode and decode two megabytes of data. To become useful in a widespread way, the price per megabyte needs to plummet. Even advocates concede this point. "Of course it is expensive," Zhirnov says. "But look how much magnetic storage cost in the 1980s. What you store today in your iPhone for virtually nothing would cost many millions of dollars in 1982." There's reason to think the price will continue to fall. Genome readers are improving, getting cheaper, faster, and smaller, and genome sequencing becomes cheaper every year, too. Picture it: tiny specks of inert DNA made from silicon or another material, stored in cool, dark, dry areas, preserved for all time.
"It just takes a few minutes to double a sample. A few more minutes, you double it again. Very quickly, you have thousands or millions of new copies."
Plus, DNA has another advantage over more traditional forms of storage: It's very easy to reproduce. "If you want a second copy of a hard disk drive, you need components for a disk drive, hook both drives up to a computer, and copy. That's a pain," Nick Goldman, a researcher at the European Bioinformatics Institute, says. "DNA, once you have that first sample, it's a process that is absolutely routine in thousands of laboratories around the world to multiply that using polymerase chain reaction [which uses temperature changes or other processes]. It just takes a few minutes to double a sample. A few more minutes, you double it again. Very quickly, you have thousands or millions of new copies."
This ability to duplicate quickly and easily is a positive trait. But, of course, there's also the potential for danger. Does encoding on DNA, the very basis for life, present ethical issues? Could it get out of control and fundamentally alter life as we know it?
The chance is there, but it's remote. The first reason is that storage could be done with only two base pairs, which would serve as replacements for the 0 and 1 digits that make up all digital data. While doing so would decrease the possible density of the storage, it would virtually eliminate the risk that the sequences would be compatible with life.
But even if scientists and researchers choose to use four base pairs, other safeguards are in place that will prevent trouble. According to Ceze, the computer science professor, the snippets of DNA that they write are very short, around 150 nucleotides. This includes the title, the information that's being encoded, and tags to help organize where the snippet should fall in the larger sequence. Furthermore, they generally avoid repeated letters, which dramatically reduces the chance that a protein could be synthesized from the snippet.
"In the future, we'll know enough about someone from a sample of their DNA that we could make a specific poison. That's the danger, not those of us who want to encode DNA for storage."
Inevitably, some DNA will get spilt. "But it's so unlikely that anything that gets created for storage would have a biological interpretation that could interfere with the mechanisms going on in a living organism that it doesn't worry me in the slightest," Goldman says. "We're not of concern for the people who are worried about the ethical issues of synthetic DNA. They are much more concerned about people deliberately engineering anthrax. In the future, we'll know enough about someone from a sample of their DNA that we could make a specific poison. That's the danger, not those of us who want to encode DNA for storage."
In the end, the reality of and risks surrounding encoding on DNA are the same as any scientific advancement: It's another system that is vulnerable to people with bad intentions but not one that is inherently unethical.
"Every human action has some ethical implications," Zhirnov says. "I can use a hammer to build a house or I can use it to harm another person. I don't see why DNA is in any way more or less ethical."
If that house can store all the knowledge in human history, it's worth learning how to build it.
Editor's Note: In response to readers' comments that silicon is one of the earth's most abundant materials, we reached back out to our source, Dr. Victor Zhirnov. He stands by his statement about a coming shortage of silicon, citing this research. The silicon oxide found in beach sand is unsuitable for semiconductors, he says, because the cost of purifying it would be prohibitive. For use in circuit-making, silicon must be refined to a purity of 99.9999999 percent. So the process begins by mining for pure quartz, which can only be found in relatively few places around the world.
Jamie Rettinger with his now fiance Amie Purnel-Davis, who helped him through the clinical trial.
Jamie Rettinger was still in his thirties when he first noticed a tiny streak of brown running through the thumbnail of his right hand. It slowly grew wider and the skin underneath began to deteriorate before he went to a local dermatologist in 2013. The doctor thought it was a wart and tried scooping it out, treating the affected area for three years before finally removing the nail bed and sending it off to a pathology lab for analysis.
"I have some bad news for you; what we removed was a five-millimeter melanoma, a cancerous tumor that often spreads," Jamie recalls being told on his return visit. "I'd never heard of cancer coming through a thumbnail," he says. None of his doctors had ever mentioned it either. "I just thought I was being treated for a wart." But nothing was healing and it continued to bleed.
A few months later a surgeon amputated the top half of his thumb. Lymph node biopsy tested negative for spread of the cancer and when the bandages finally came off, Jamie thought his medical issues were resolved.
Melanoma is the deadliest form of skin cancer. About 85,000 people are diagnosed with it each year in the U.S. and more than 8,000 die of the cancer when it spreads to other parts of the body, according to the Centers for Disease Control and Prevention (CDC).
There are two peaks in diagnosis of melanoma; one is in younger women ages 30-40 and often is tied to past use of tanning beds; the second is older men 60+ and is related to outdoor activity from farming to sports. Light-skinned people have a twenty-times greater risk of melanoma than do people with dark skin.
"When I graduated from medical school, in 2005, melanoma was a death sentence" --Diwakar Davar.
Jamie had a follow up PET scan about six months after his surgery. A suspicious spot on his lung led to a biopsy that came back positive for melanoma. The cancer had spread. Treatment with a monoclonal antibody (nivolumab/Opdivo®) didn't prove effective and he was referred to the UPMC Hillman Cancer Center in Pittsburgh, a four-hour drive from his home in western Ohio.
An alternative monoclonal antibody treatment brought on such bad side effects, diarrhea as often as 15 times a day, that it took more than a week of hospitalization to stabilize his condition. The only options left were experimental approaches in clinical trials.
Early research
"When I graduated from medical school, in 2005, melanoma was a death sentence" with a cure rate in the single digits, says Diwakar Davar, 39, an oncologist at UPMC Hillman Cancer Center who specializes in skin cancer. That began to change in 2010 with introduction of the first immunotherapies, monoclonal antibodies, to treat cancer. The antibodies attach to PD-1, a receptor on the surface of T cells of the immune system and on cancer cells. Antibody treatment boosted the melanoma cure rate to about 30 percent. The search was on to understand why some people responded to these drugs and others did not.
At the same time, there was a growing understanding of the role that bacteria in the gut, the gut microbiome, plays in helping to train and maintain the function of the body's various immune cells. Perhaps the bacteria also plays a role in shaping the immune response to cancer therapy.
One clue came from genetically identical mice. Animals ordered from different suppliers sometimes responded differently to the experiments being performed. That difference was traced to different compositions of their gut microbiome; transferring the microbiome from one animal to another in a process known as fecal transplant (FMT) could change their responses to disease or treatment.
When researchers looked at humans, they found that the patients who responded well to immunotherapies had a gut microbiome that looked like healthy normal folks, but patients who didn't respond had missing or reduced strains of bacteria.
Davar and his team knew that FMT had a very successful cure rate in treating the gut dysbiosis of Clostridioides difficile, a persistant intestinal infection, and they wondered if a fecal transplant from a patient who had responded well to cancer immunotherapy treatment might improve the cure rate of patients who did not originally respond to immunotherapies for melanoma.
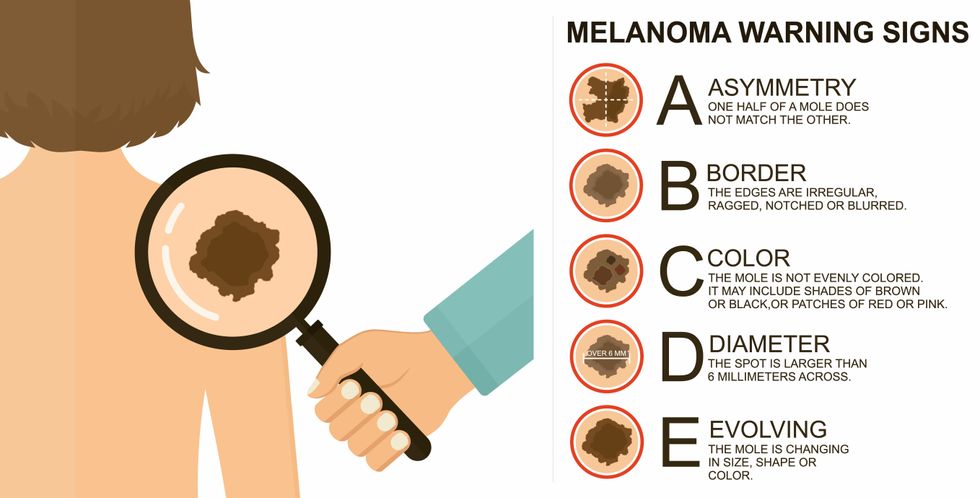
The ABCDE of melanoma detection
Adobe Stock
Clinical trial
"It was pretty weird, I was totally blasted away. Who had thought of this?" Jamie first thought when the hypothesis was explained to him. But Davar's explanation that the procedure might restore some of the beneficial bacterial his gut was lacking, convinced him to try. He quickly signed on in October 2018 to be the first person in the clinical trial.
Fecal donations go through the same safety procedures of screening for and inactivating diseases that are used in processing blood donations to make them safe for transfusion. The procedure itself uses a standard hollow colonoscope designed to screen for colon cancer and remove polyps. The transplant is inserted through the center of the flexible tube.
Most patients are sedated for procedures that use a colonoscope but Jamie doesn't respond to those drugs: "You can't knock me out. I was watching them on the TV going up my own butt. It was kind of unreal at that point," he says. "There were about twelve people in there watching because no one had seen this done before."
A test two weeks after the procedure showed that the FMT had engrafted and the once-missing bacteria were thriving in his gut. More importantly, his body was responding to another monoclonal antibody (pembrolizumab/Keytruda®) and signs of melanoma began to shrink. Every three months he made the four-hour drive from home to Pittsburgh for six rounds of treatment with the antibody drug.
"We were very, very lucky that the first patient had a great response," says Davar. "It allowed us to believe that even though we failed with the next six, we were on the right track. We just needed to tweak the [fecal] cocktail a little better" and enroll patients in the study who had less aggressive tumor growth and were likely to live long enough to complete the extensive rounds of therapy. Six of 15 patients responded positively in the pilot clinical trial that was published in the journal Science.
Davar believes they are beginning to understand the biological mechanisms of why some patients initially do not respond to immunotherapy but later can with a FMT. It is tied to the background level of inflammation produced by the interaction between the microbiome and the immune system. That paper is not yet published.
Surviving cancer
It has been almost a year since the last in his series of cancer treatments and Jamie has no measurable disease. He is cautiously optimistic that his cancer is not simply in remission but is gone for good. "I'm still scared every time I get my scans, because you don't know whether it is going to come back or not. And to realize that it is something that is totally out of my control."
"It was hard for me to regain trust" after being misdiagnosed and mistreated by several doctors he says. But his experience at Hillman helped to restore that trust "because they were interested in me, not just fixing the problem."
He is grateful for the support provided by family and friends over the last eight years. After a pause and a sigh, the ruggedly built 47-year-old says, "If everyone else was dead in my family, I probably wouldn't have been able to do it."
"I never hesitated to ask a question and I never hesitated to get a second opinion." But Jamie acknowledges the experience has made him more aware of the need for regular preventive medical care and a primary care physician. That person might have caught his melanoma at an earlier stage when it was easier to treat.
Davar continues to work on clinical studies to optimize this treatment approach. Perhaps down the road, screening the microbiome will be standard for melanoma and other cancers prior to using immunotherapies, and the FMT will be as simple as swallowing a handful of freeze-dried capsules off the shelf rather than through a colonoscopy. Earlier this year, the Food and Drug Administration approved the first oral fecal microbiota product for C. difficile, hopefully paving the way for more.
An older version of this hit article was first published on May 18, 2021
All organisms can repair damaged tissue, but none do it better than salamanders and newts. A surprising area of science could tell us how they manage this feat - and perhaps even help us develop a similar ability.
All organisms have the capacity to repair or regenerate tissue damage. None can do it better than salamanders or newts, which can regenerate an entire severed limb.
That feat has amazed and delighted man from the dawn of time and led to endless attempts to understand how it happens – and whether we can control it for our own purposes. An exciting new clue toward that understanding has come from a surprising source: research on the decline of cells, called cellular senescence.
Senescence is the last stage in the life of a cell. Whereas some cells simply break up or wither and die off, others transition into a zombie-like state where they can no longer divide. In this liminal phase, the cell still pumps out many different molecules that can affect its neighbors and cause low grade inflammation. Senescence is associated with many of the declining biological functions that characterize aging, such as inflammation and genomic instability.
Oddly enough, newts are one of the few species that do not accumulate senescent cells as they age, according to research over several years by Maximina Yun. A research group leader at the Center for Regenerative Therapies Dresden and the Max Planck Institute of Molecular and Cell Biology and Genetics, in Dresden, Germany, Yun discovered that senescent cells were induced at some stages of regeneration of the salamander limb, “and then, as the regeneration progresses, they disappeared, they were eliminated by the immune system,” she says. “They were present at particular times and then they disappeared.”
Senescent cells added to the edges of the wound helped the healthy muscle cells to “dedifferentiate,” essentially turning back the developmental clock of those cells into more primitive states.
Previous research on senescence in aging had suggested, logically enough, that applying those cells to the stump of a newly severed salamander limb would slow or even stop its regeneration. But Yun stood that idea on its head. She theorized that senescent cells might also play a role in newt limb regeneration, and she tested it by both adding and removing senescent cells from her animals. It turned out she was right, as the newt limbs grew back faster than normal when more senescent cells were included.
Senescent cells added to the edges of the wound helped the healthy muscle cells to “dedifferentiate,” essentially turning back the developmental clock of those cells into more primitive states, which could then be turned into progenitors, a cell type in between stem cells and specialized cells, needed to regrow the muscle tissue of the missing limb. “We think that this ability to dedifferentiate is intrinsically a big part of why salamanders can regenerate all these very complex structures, which other organisms cannot,” she explains.
Yun sees regeneration as a two part problem. First, the cells must be able to sense that their neighbors from the lost limb are not there anymore. Second, they need to be able to produce the intermediary progenitors for regeneration, , to form what is missing. “Molecularly, that must be encoded like a 3D map,” she says, otherwise the new tissue might grow back as a blob, or liver, or fin instead of a limb.
Wound healing
Another recent study, this time at the Mayo Clinic, provides evidence supporting the role of senescent cells in regeneration. Looking closely at molecules that send information between cells in the wound of a mouse, the researchers found that senescent cells appeared near the start of the healing process and then disappeared as healing progressed. In contrast, persistent senescent cells were the hallmark of a chronic wound that did not heal properly. The function and significance of senescence cells depended on both the timing and the context of their environment.
The paper suggests that senescent cells are not all the same. That has become clearer as researchers have been able to identify protein markers on the surface of some senescent cells. The patterns of these proteins differ for some senescent cells compared to others. In biology, such physical differences suggest functional differences, so it is becoming increasingly likely there are subsets of senescent cells with differing functions that have not yet been identified.
There are disagreements within the research community as to whether newts have acquired their regenerative capacity through a unique evolutionary change, or if other animals, including humans, retain this capacity buried somewhere in their genes.
Scientists initially thought that senescent cells couldn’t play a role in regeneration because they could no longer reproduce, says Anthony Atala, a practicing surgeon and bioengineer who leads the Wake Forest Institute for Regenerative Medicine in North Carolina. But Yun’s study points in the other direction. “What this paper shows clearly is that these cells have the potential to be involved in tissue regeneration [in newts]. The question becomes, will these cells be able to do the same in humans.”
As our knowledge of senescent cells increases, Atala thinks we need to embrace a new analogy to help understand them: humans in retirement. They “have acquired a lot of wisdom throughout their whole life and they can help younger people and mentor them to grow to their full potential. We're seeing the same thing with these cells,” he says. They are no longer putting energy into their own reproduction, but the signaling molecules they secrete “can help other cells around them to regenerate.”
There are disagreements within the research community as to whether newts have acquired their regenerative capacity through a unique evolutionary change, or if other animals, including humans, retain this capacity buried somewhere in their genes. If so, it seems that our genes are unable to express this ability, perhaps as part of a tradeoff in acquiring other traits. It is a fertile area of research.
Dedifferentiation is likely to become an important process in the field of regenerative medicine. One extreme example: a lab has been able to turn back the clock and reprogram adult male skin cells into female eggs, a potential milestone in reproductive health. It will be more difficult to control just how far back one wishes to go in the cell's dedifferentiation – part way or all the way back into a stem cell – and then direct it down a different developmental pathway. Yun is optimistic we can learn these tricks from newts.
Senolytics
A growing field of research is using drugs called senolytics to remove senescent cells and slow or even reverse disease of aging.
“Senolytics are great, but senolytics target different types of senescence,” Yun says. “If senescent cells have positive effects in the context of regeneration, of wound healing, then maybe at the beginning of the regeneration process, you may not want to take them out for a little while.”
“If you look at pretty much all biological systems, too little or too much of something can be bad, you have to be in that central zone” and at the proper time, says Atala. “That's true for proteins, sugars, and the drugs that you take. I think the same thing is true for these cells. Why would they be different?”
Our growing understanding that senescence is not a single thing but a variety of things likely means that effective senolytic drugs will not resemble a single sledge hammer but more a carefully manipulated scalpel where some types of senescent cells are removed while others are added. Combinations and timing could be crucial, meaning the difference between regenerating healthy tissue, a scar, or worse.

