Can You Trust Your Gut for Food Advice?
Kira Peikoff was the editor-in-chief of Leaps.org from 2017 to 2021. As a journalist, her work has appeared in The New York Times, Newsweek, Nautilus, Popular Mechanics, The New York Academy of Sciences, and other outlets. She is also the author of four suspense novels that explore controversial issues arising from scientific innovation: Living Proof, No Time to Die, Die Again Tomorrow, and Mother Knows Best. Peikoff holds a B.A. in Journalism from New York University and an M.S. in Bioethics from Columbia University. She lives in New Jersey with her husband and two young sons. Follow her on Twitter @KiraPeikoff.

Which foods are actually healthy for your individual gut microbiome? Several companies are offering personalized dietary guidance based on your test results, but their answers in one experiment turned up with some conflicting advice.
I recently got on the scale to weigh myself, thinking I've got to eat better. With so many trendy diets today claiming to improve health, from Keto to Paleo to Whole30, it can be confusing to figure out what we should and shouldn't eat for optimal nutrition.
A number of companies are now selling the concept of "personalized" nutrition based on the genetic makeup of your individual gut bugs.
My next thought was: I've got to lose a few pounds.
Consider a weird factoid: In addition to my fat, skin, bone and muscle, I'm carrying around two or three pounds of straight-up bacteria. Like you, I am the host to trillions of micro-organisms that live in my gut and are collectively known as my microbiome. An explosion of research has occurred in the last decade to try to understand exactly how these microbial populations, which are unique to each of us, may influence our overall health and potentially even our brains and behavior.
Lots of mysteries still remain, but it is established that these "bugs" are crucial to keeping our body running smoothly, performing functions like stimulating the immune system, synthesizing important vitamins, and aiding digestion. The field of microbiome science is evolving rapidly, and a number of companies are now selling the concept of "personalized" nutrition based on the genetic makeup of your individual gut bugs. The two leading players are Viome and DayTwo, but the landscape includes the newly launched startup Onegevity Health and others like Thryve, which offers customized probiotic supplements in addition to dietary recommendations.
The idea has immediate appeal – if science could tell you exactly what to make for lunch and what to avoid, you could forget about the fad diets and go with your own bespoke food pyramid. Wondering if the promise might be too good to be true, I decided to perform my own experiment.
Last fall, I sent the identical fecal sample to both Viome (I paid $425, but the price has since dropped to $299) and DayTwo ($349). A couple of months later, both reports finally arrived, and I eagerly opened each app to compare their recommendations.
First, I examined my results from Viome, which was founded in 2016 in Cupertino, Calif., and declares without irony on its website that "conflicting food advice is now obsolete."
I learned I have "average" metabolic fitness and "average" inflammatory activity in my gut, which are scores that the company defines based on a proprietary algorithm. But I have "low" microbial richness, with only 62 active species of bacteria identified in my sample, compared with the mean of 157 in their test population. I also received a list of the specific species in my gut, with names like Lactococcus and Romboutsia.
But none of it meant anything to me without actionable food advice, so I clicked through to the Recommendations page and found a list of My Superfoods (cranberry, garlic, kale, salmon, turmeric, watermelon, and bone broth) and My Foods to Avoid (chickpeas, kombucha, lentils, and rice noodles). There was also a searchable database of many foods that had been categorized for me, like "bell pepper; minimize" and "beef; enjoy."
"I just don't think sufficient data is yet available to make reliable personalized dietary recommendations based on one's microbiome."
Next, I looked at my results from DayTwo, which was founded in 2015 from research out of the Weizmann Institute of Science in Israel, and whose pitch to consumers is, "Blood sugar made easy. The algorithm diet personalized to you."
This app had some notable differences. There was no result about my metabolic fitness, microbial richness, or list of the species in my sample. There was also no list of superfoods or foods to avoid. Instead, the app encouraged me to build a meal by searching for foods in their database and combining them in beneficial ways for my blood sugar. Two slices of whole wheat bread received a score of 2.7 out of 10 ("Avoid"), but if combined with one cup of large curd cottage cheese, the score improved to 6.8 ("Limit"), and if I added two hard-boiled eggs, the score went up to 7.5 ("Good").
Perusing my list of foods with "Excellent" scores, I noticed some troubling conflicts with the other app. Lentils, which had been a no-no according to Viome, received high marks from DayTwo. Ditto for Kombucha. My purported superfood of cranberry received low marks. Almonds got an almost perfect score (9.7) while Viome told me to minimize them. I found similarly contradictory advice for foods I regularly eat, including navel oranges, peanuts, pork, and beets.
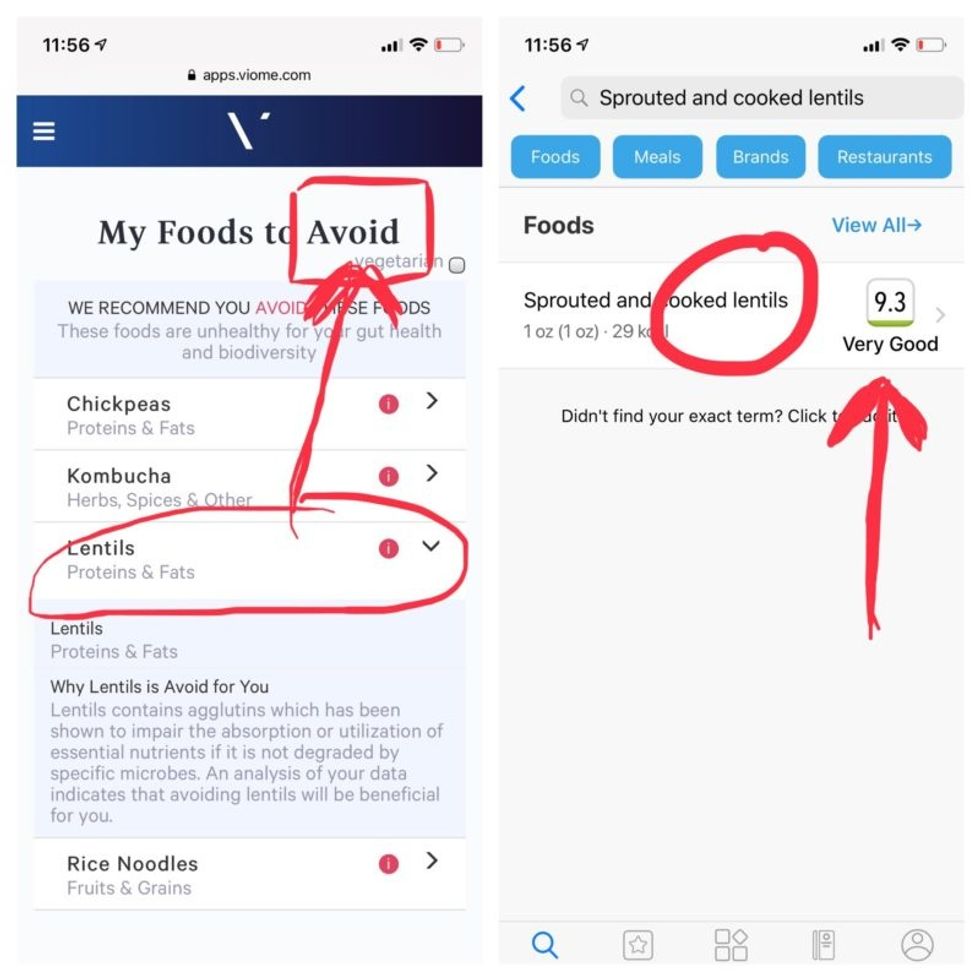
Contradictory dietary guidance that Kira Peikoff received from Viome (left) and DayTwo from an identical sample.
To be sure, there was some overlap. Both apps agreed on rice noodles (bad), chickpeas (bad), honey (bad), carrots (good), and avocado (good), among other foods.
But still, I was left scratching my head. Which set of recommendations should I trust, if either? And what did my results mean for the accuracy of this nascent field?
I called a couple of experts to find out.
"I have worked on the microbiome and nutrition for the last 20 years and I would be absolutely incapable of finding you evidence in the scientific literature that lentils have a detrimental effect based on the microbiome," said Dr. Jens Walter, an Associate Professor and chair for Nutrition, Microbes, and Gastrointestinal Health at the University of Alberta. "I just don't think sufficient data is yet available to make reliable personalized dietary recommendations based on one's microbiome. And even if they would have proprietary algorithms, at least one of them is not doing it right."
There is definite potential for personalized nutrition based on the microbiome, he said, but first, predictive models must be built and standardized, then linked to clinical endpoints, and tested in a large sample of healthy volunteers in order to enable extrapolations for the general population.
"It is mindboggling what you would need to do to make this work," he observed. "There are probably hundreds of relevant dietary compounds, then the microbiome has at least a hundred relevant species with a hundred or more relevant genes each, then you'd have to put all this together with relevant clinical outcomes. And there's a hundred-fold variation in that information between individuals."
However, Walter did acknowledge that the companies might be basing their algorithms on proprietary data that could potentially connect all the dots. I reached out to them to find out.
Amir Golan, the Chief Commercial Officer of DayTwo, told me, "It's important to emphasize this is a prediction, as the microbiome field is in a very early stage of research." But he added, "I believe we are the only company that has very solid science published in top journals and we can bring very actionable evidence and benefit to our uses."
He was referring to pioneering work out of the Weizmann Institute that was published in 2015 in the journal Cell, which logged the glycemic responses of 800 people in response to nearly 50,000 meals; adding information about the subjects' microbiomes enabled more accurate glycemic response predictions. Since then, Golan said, additional trials have been conducted, most recently with the Mayo Clinic, to duplicate the results, and other studies are ongoing whose results have not yet been published.
He also pointed out that the microbiome was merely one component that goes into building a client's profile, in addition to medical records, including blood glucose levels. (I provided my HbA1c levels, a measure of average blood sugar over the previous several months.)
"We are not saying we want to improve your gut microbiome. We provide a dynamic tool to help guide what you should eat to control your blood sugar and think about combinations," he said. "If you eat one thing, or with another, it will affect you in a different way."
Viome acknowledged that the two companies are taking very different approaches.
"DayTwo is primarily focused on the glycemic response," Naveen Jain, the CEO, told me. "If you can only eat butter for rest of your life, you will have no glycemic response but will probably die of a heart attack." He laughed. "Whereas we came from very different angle – what is happening inside the gut at a microbial level? When you eat food like spinach, how will that be metabolized in the gut? Will it produce the nutrients you need or cause inflammation?"
He said his team studied 1000 people who were on continuous glucose monitoring and fed them 45,000 meals, then built a proprietary data prediction model, looking at which microbes existed and how they actively broke down the food.
Jain pointed out that DayTwo sequences the DNA of the microbes, while Viome sequences the RNA – the active expression of DNA. That difference, in his opinion, is key to making accurate predictions.
"DNA is extremely stable, so when you eat any food and measure the DNA [in a fecal sample], you get all these false positives--you get DNA from plant food and meat, and you have no idea if those organisms are dead and simply transient, or actually exist. With RNA, you see what is actually alive in the gut."
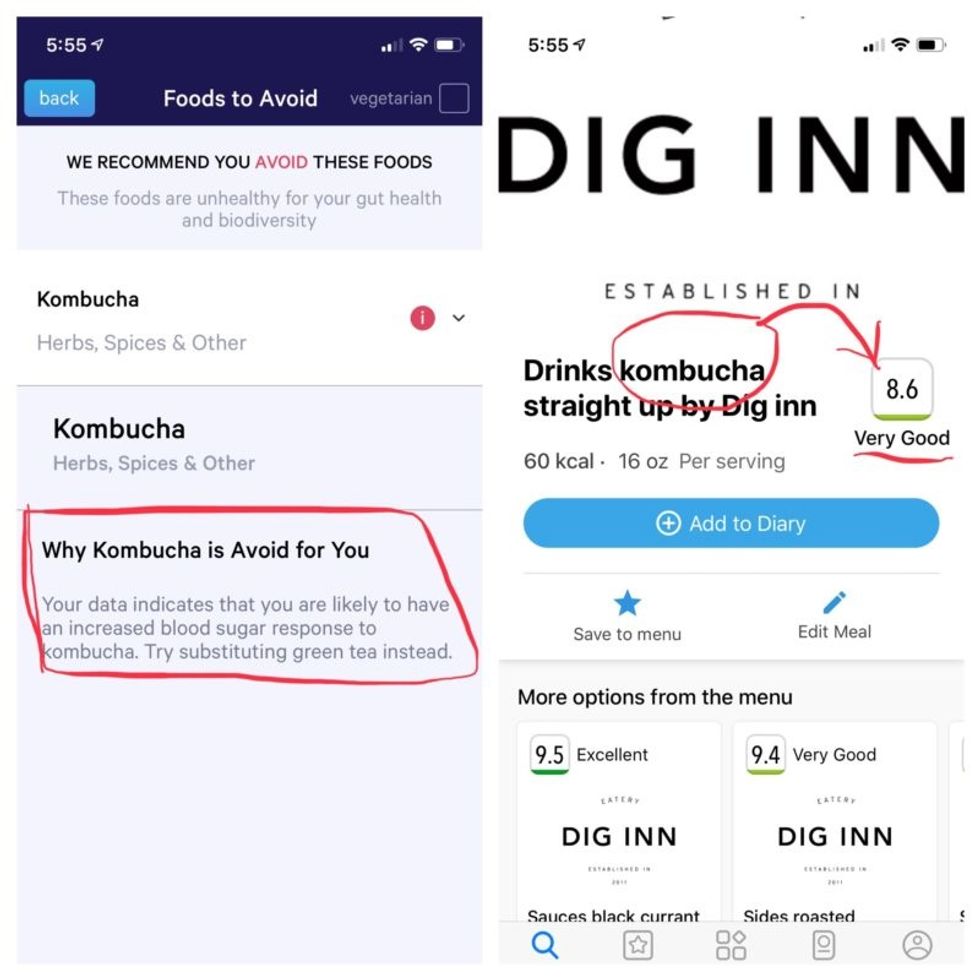
More contradictory food advice from Viome (left) and DayTwo.
Note that controversy exists over how it is possible with a fecal sample to effectively measure RNA, which degrades within minutes, though Jain said that his company has the technology to keep RNA stable for fourteen days.
Viome's approach, Jain maintains, is 90 percent accurate, based on as-yet unpublished data; a patent was filed just last week. DayTwo's approach is 66 percent accurate according to the latest published research.
Natasha Haskey, a registered dietician and doctoral student conducting research in the field of microbiome science and nutrition, is skeptical of both companies. "We can make broad statements, like eat more fruits and vegetables and fiber, but when it comes to specific foods, the science is just not there yet," she said. "I think there is a future, and we will be doing that someday, but not yet. Maybe we will be closer in ten years."
Professor Walter wholeheartedly agrees with Haskey, and suggested that if people want to eat a gut-healthy diet, they should focus on beneficial oils, fruits and vegetables, fish, a variety of whole grains, poultry and beans, and limit red meat and cheese, as well as avoid processed meats.
"These services are far over the tips of their science skis," Arthur Caplan, the founding head of New York University's Division of Medical Ethics, said in an email. "We simply don't know enough about the gut microbiome, its fluctuations and variability from person to person to support general [direct-to-consumer] testing. This is simply premature. We need standards for accuracy, specificity, and sensitivity, plus mandatory competent counseling for all such testing. They don't exist. Neither should DTC testing—yet."
Meanwhile, it's time for lunch. I close out my Viome and DayTwo apps and head to the kitchen to prepare a peanut butter sandwich. My gut tells me I'll be just fine.
Kira Peikoff was the editor-in-chief of Leaps.org from 2017 to 2021. As a journalist, her work has appeared in The New York Times, Newsweek, Nautilus, Popular Mechanics, The New York Academy of Sciences, and other outlets. She is also the author of four suspense novels that explore controversial issues arising from scientific innovation: Living Proof, No Time to Die, Die Again Tomorrow, and Mother Knows Best. Peikoff holds a B.A. in Journalism from New York University and an M.S. in Bioethics from Columbia University. She lives in New Jersey with her husband and two young sons. Follow her on Twitter @KiraPeikoff.
The U.S. must fund more biotech innovation – or other countries will catch up faster than you think
In the coming years, U.S. market share in biotech will decline unless the federal government makes investments to improve the quality and quantity of U.S. research, writes the author.
The U.S. has approximately 58 percent of the market share in the biotech sector, followed by China with 11 percent. However, this market share is the result of several years of previous research and development (R&D) – it is a present picture of what happened in the past. In the future, this market share will decline unless the federal government makes investments to improve the quality and quantity of U.S. research in biotech.
The effectiveness of current R&D can be evaluated in a variety of ways such as monies invested and the number of patents filed. According to the UNESCO Institute for Statistics, the U.S. spends approximately 2.7 percent of GDP on R&D ($476,459.0M), whereas China spends 2 percent ($346,266.3M). However, investment levels do not necessarily translate into goods that end up contributing to innovation.
Patents are a better indication of innovation. The biotech industry relies on patents to protect their investments, making patenting a key tool in the process of translating scientific discoveries that can ultimately benefit patients. In 2020, China filed 1,497,159 patents, a 6.9 percent increase in growth rate. In contrast, the U.S. filed 597,172, a 3.9 percent decline. When it comes to patents filed, China has approximately 45 percent of the world share compared to 18 percent for the U.S.
So how did we get here? The nature of science in academia allows scientists to specialize by dedicating several years to advance discovery research and develop new inventions that can then be licensed by biotech companies. This makes academic science critical to innovation in the U.S. and abroad.
Academic scientists rely on government and foundation grants to pay for R&D, which includes salaries for faculty, investigators and trainees, as well as monies for infrastructure, support personnel and research supplies. Of particular interest to academic scientists to cover these costs is government support such as Research Project Grants, also known as R01 grants, the oldest grant mechanism from the National Institutes of Health. Unfortunately, this funding mechanism is extremely competitive, as applications have a success rate of only about 20 percent. To maximize the chances of getting funded, investigators tend to limit the innovation of their applications, since a project that seems overambitious is discouraged by grant reviewers.
Considering the difficulty in obtaining funding, the limited number of opportunities for scientists to become independent investigators capable of leading their own scientific projects, and the salaries available to pay for scientists with a doctoral degree, it is not surprising that the U.S. is progressively losing its workforce for innovation.
This approach affects the future success of the R&D enterprise in the U.S. Pursuing less innovative work tends to produce scientific results that are more obvious than groundbreaking, and when a discovery is obvious, it cannot be patented, resulting in fewer inventions that go on to benefit patients. Even though there are governmental funding options available for scientists in academia focused on more groundbreaking and translational projects, those options are less coveted by academic scientists who are trying to obtain tenure and long-term funding to cover salaries and other associated laboratory expenses. Therefore, since only a small percent of projects gets funded, the likelihood of scientists interested in pursuing academic science or even research in general keeps declining over time.
Efforts to raise the number of individuals who pursue a scientific education are paying off. However, the number of job openings for those trainees to carry out independent scientific research once they graduate has proved harder to increase. These limitations are not just in the number of faculty openings to pursue academic science, which are in part related to grant funding, but also the low salary available to pay those scientists after they obtain their doctoral degree, which ranges from $53,000 to $65,000, depending on years of experience.
Thus, considering the difficulty in obtaining funding, the limited number of opportunities for scientists to become independent investigators capable of leading their own scientific projects, and the salaries available to pay for scientists with a doctoral degree, it is not surprising that the U.S. is progressively losing its workforce for innovation, which results in fewer patents filed.
Perhaps instead of encouraging scientists to propose less innovative projects in order to increase their chances of getting grants, the U.S. government should give serious consideration to funding investigators for their potential for success -- or the success they have already achieved in contributing to the advancement of science. Such a funding approach should be tiered depending on career stage or years of experience, considering that 42 years old is the median age at which the first R01 is obtained. This suggests that after finishing their training, scientists spend 10 years before they establish themselves as independent academic investigators capable of having the appropriate funds to train the next generation of scientists who will help the U.S. maintain or even expand its market share in the biotech industry for years to come. Patenting should be given more weight as part of the academic endeavor for promotion purposes, or governmental investment in research funding should be increased to support more than just 20 percent of projects.
Remaining at the forefront of biotech innovation will give us the opportunity to not just generate more jobs, but it will also allow us to attract the brightest scientists from all over the world. This talented workforce will go on to train future U.S. scientists and will improve our standard of living by giving us the opportunity to produce the next generation of therapies intended to improve human health.
This problem cannot rely on just one solution, but what is certain is that unless there are more creative changes in funding approaches for scientists in academia, eventually we may be saying “remember when the U.S. was at the forefront of biotech innovation?”
New gene therapy helps patients with rare disease. One mother wouldn't have it any other way.
A biotech in Cambridge, Mass., is targeting a rare disease called cystinosis with gene therapy. It's been effective for five patients in a clinical trial that's still underway.
Three years ago, Jordan Janz of Consort, Alberta, knew his gene therapy treatment for cystinosis was working when his hair started to darken. Pigmentation or melanin production is just one part of the body damaged by cystinosis.
“When you have cystinosis, you’re either a redhead or a blonde, and you are very pale,” attests Janz, 23, who was diagnosed with the disease just eight months after he was born. “After I got my new stem cells, my hair came back dark, dirty blonde, then it lightened a little bit, but before it was white blonde, almost bleach blonde.”
According to Cystinosis United, about 500 to 600 people have the rare genetic disease in the U.S.; an estimated 20 new cases are diagnosed each year.
Located in Cambridge, Mass., AVROBIO is a gene therapy company that targets cystinosis and other lysosomal storage disorders, in which toxic materials build up in the cells. Janz is one of five patients in AVROBIO’s ongoing Phase 1/2 clinical trial of a gene therapy for cystinosis called AVR-RD-04.
Recently, AVROBIO compiled positive clinical data from this first and only gene therapy trial for the disease. The data show the potential of the therapy to genetically modify the patients’ own hematopoietic stem cells—a certain type of cell that’s capable of developing into all different types of blood cells—to express the functional protein they are deficient in. It stabilizes or reduces the impact of cystinosis on multiple tissues with a single dose.
Medical researchers have found that more than 80 different mutations to a gene called CTNS are responsible for causing cystinosis. The most common mutation results in a deficiency of the protein cystinosin. That protein functions as a transporter that regulates a lot metabolic processes in the cells.
“One of the first things we see in patients clinically is an accumulation of a particular amino acid called cystine, which grows toxic cystine crystals in the cells that cause serious complications,” explains Essra Rihda, chief medical officer for AVROBIO. “That happens in the cells across the tissues and organs of the body, so the disease affects many parts of the body.”

Jordan Janz, 23, meets Stephanie Cherqui, the principal investigator of his gene therapy trial, before the trial started in 2019.
Jordan Janz
According to Rihda, although cystinosis can occur in kids and adults, the most severe form of the disease affects infants and makes up about 95 percent of overall cases. Children typically appear healthy at birth, but around six to 18 months, they start to present for medical attention with failure to thrive.
Additionally, infants with cystinosis often urinate frequently, a sign of polyuria, and they are thirsty all the time, since the disease usually starts in the kidneys. Many develop chronic kidney disease that ultimately progresses to the point where the kidney no longer supports the body’s needs. At that stage, dialysis is required and then a transplant. From there the disease spreads to many other organs, including the eyes, muscles, heart, nervous system, etc.
“The gene for cystinosis is expressed in every single tissue we have, and the accumulation of this toxic buildup alters all of the organs of the patient, so little by little all of the organs start to fail,” says Stephanie Cherqui, principal investigator of Cherqui Lab, which is part of UC San Diego’s Department of Pediatrics.
Since the 1950s, a drug called cysteamine showed some therapeutic effect on cystinosis. It was approved by the FDA in 1994 to prevent damage that may be caused by the buildup of cystine crystals in organs. Prior to FDA approval, Cherqui says, children were dying of the disease before they were ten-years-old or after a kidney transplant. By taking oral cysteamine, they can live from 20 to 50 years longer. But it’s a challenging drug because it has to be taken every 6 or 12 hours, and there are serious gastric side effects such as nausea and diarrhea.
“With all of the complications they develop, the typical patient takes 40 to 60 pills a day around the clock,” Cherqui says. “They literally have a suitcase of medications they have to carry everywhere, and all of those medications don’t stop the progression of the disease, and they still die from it.”
Cherqui has been a proponent of gene therapy to treat children’s disorders since studying cystinosis while earning her doctorate in 2002. Today, her lab focuses on developing stem cell and gene therapy strategies for degenerative, hereditary disorders such as cystinosis that affect multiple systems of the body. “Because cystinosis expresses in every tissue in the body, I decided to use the blood-forming stem cells that we have in our bone marrow,” she explains. “These cells can migrate to anywhere in the body where the person has an injury from the disease.”
AVROBIO’s hematopoietic stem cell gene therapy approach collects stem cells from the patient’s bone marrow. They then genetically modify the stem cells to give the patient a copy of the healthy CTNS gene, which the person either doesn’t have or it’s defective.
The patient first undergoes apheresis, a medical procedure in which their blood is passed through an apparatus that separates out the diseased stem cells, and a process called conditioning is used to help eliminate the damaged cells so they can be replaced by the infusion of the patient’s genetically modified stem cells. Once they become engrafted into the patient’s bone marrow, they reproduce into a lot of daughter cells, and all of those daughter cells contain the CTNS gene. Those cells are able to express the healthy, functional, active protein throughout the body to correct the metabolic problem caused by cystinosis.
“What we’re seeing in the adult patients who have been dosed to date is the consistent and sustained engraftment of our genetically modified cells, 17 to 27 months post-gene therapy, so that’s very encouraging and positive,” says Rihda, the chief medical officer at AVROBIO.
When Janz was 11-years-old, his mother got him enrolled in the trial of a new form of cysteamine that would only need to be taken every 12 hours instead of every six. Two years later, she made sure he was the first person on the list for Cherqui’s current stem cell gene therapy trial.
AVROBIO researchers have also confirmed stabilization or improvement in motor coordination and visual perception in the trial participants, suggesting a potential impact on the neuropathology of the disease. Data from five dosed patients show strong safety and tolerability as well as reduced accumulation of cystine crystals in cells across multiple tissues in the first three patients. None of the five patients need to take oral cysteamine.
Janz’s mother, Barb Kulyk, whom he credits with always making him take his medications and keeping him hydrated, had been following Cherqui’s research since his early childhood. When Janz was 11-years-old, she got him enrolled in the trial of a new form of cysteamine that would only need to be taken every 12 hours instead of every six. When he was 17, the FDA approved that drug. Two years later, his mother made sure he was the first person on the list for Cherqui’s current stem cell gene therapy trial. He received his new stem cells on October 7th, 2019, went home in January 2020, and returned to working full time in February.

Jordan Janz, pictured here with his girlfriend, has a new lease on life, plus a new hair color.
Jordan Janz
He notes that his energy level is significantly better, and his mother has noticed much improvement in him and his daily functioning: He rarely vomits or gets nauseous in the morning, and he has more color in his face as well as his hair. Although he could finish his participation at any time, he recently decided to continue in the clinical trial.
Before the trial, Janz was taking 56 pills daily. He is completely off all of those medications and only takes pills to keep his kidneys working. Because of the damage caused by cystinosis over the course of his life, he’s down to about 20 percent kidney function and will eventually need a transplant.
“Some day, though, thanks to Dr. Cherqui’s team and AVROBIO’s work, when I get a new kidney, cystinosis won’t destroy it,” he concludes.

