COVID Variants Are Like “a Thief Changing Clothes” – and Our Camera System Barely Exists

Being able to track variants of concern in real time is crucial to our ability to stay ahead of the virus.
Whether it's "natural selection" as Darwin called it, or it's "mutating" as the X-Men called it, living organisms change over time, developing thumbs or more efficient protein spikes, depending on the organism and the demands of its environment. The coronavirus that causes COVID-19, SARS-CoV-2, is not an exception, and now, after the virus has infected millions of people around the globe for more than a year, scientists are beginning to see those changes.
The notorious variants that have popped up include B.1.1.7, sometimes called the UK variant, as well as P.1 and B.1.351, which seem to have emerged in Brazil and South Africa respectively. As vaccinations are picking up pace, officials are warning that now
is not the time to become complacent or relax restrictions because the variants aren't well understood.
Some appear to be more transmissible, and deadlier, while others can evade the immune system's defenses better than earlier versions of the virus, potentially undermining the effectiveness of vaccines to some degree. Genomic surveillance, the process of sequencing the genetic code of the virus widely to observe changes and patterns, is a critical way that scientists can keep track of its evolution and work to understand how the variants might affect humans.
"It's like a thief changing clothes"
It's important to note that viruses mutate all the time. If there were funding and personnel to sequence the genome of every sample of the virus, scientists would see thousands of mutations. Not every variant deserves our attention. The vast majority of mutations are not important at all, but recognizing those that are is a crucial tool in getting and staying ahead of the virus. The work of sequencing, analyzing, observing patterns, and using public health tools as necessary is complicated and confusing to those without years of specialized training.
Jeremy Kamil, associate professor of microbiology and immunology at LSU Health Shreveport, in Louisiana, says that the variants developing are like a thief changing clothes. The thief goes in your house, steals your stuff, then leaves and puts on a different shirt and a wig, in the hopes you won't recognize them. Genomic surveillance catches the "thief" even in those different clothes.
One of the tricky things about variants is recognizing the point at which they move from interesting, to concerning at a local level, to dangerous in a larger context.
Understanding variants, both the uninteresting ones and the potentially concerning ones, gives public health officials and researchers at different levels a useful set of tools. Locally, knowing which variants are circulating in the community helps leaders know whether mask mandates and similar measures should be implemented or discontinued, or whether businesses and schools can open relatively safely.
There's more to it than observing new variants
Analysis is complex, particularly when it comes to understanding which variants are of concern. "So the question is always if a mutation becomes common, is that a random occurrence?" says Phoebe Lostroh, associate professor of molecular biology at Colorado College. "Or is the variant the result of some kind of selection because the mutation changes some property about the virus that makes it reproduce more quickly than variants of the virus that don't have that mutation? For a virus, [mutations can affect outcomes like] how much it replicates inside a person's body, how much somebody breathes it out, whether the particles that somebody might breathe in get smaller and can lead to greater transmission."
Along with all of those factors, accurate and useful genomic surveillance requires an understanding of where variants are occurring, how they are related, and an examination of why they might be prevalent.
For example, if a potentially worrisome variant appears in a community and begins to spread very quickly, it's not time to raise a public health alarm until several important questions have been answered, such as whether the variant is spreading due to specific events, or if it's happening because the mutation has allowed the virus to infect people more efficiently. Kamil offered a hypothetical scenario to explain: Imagine that a member of a community became infected and the virus mutated. That person went to church and three more people were infected, but one of them went to a karaoke bar and while singing infected 100 other people. Examining the conditions under which the virus has spread is, therefore, an essential part of untangling whether a mutation itself made the virus more transmissible or if an infected person's behaviors contributed to a local outbreak.
One of the tricky things about variants is recognizing the point at which they move from interesting, to concerning at a local level, to dangerous in a larger context. Genomic sequencing can help with that, but only when it's coordinated. When the same mutation occurs frequently, but is localized to one region, it's a concern, but when the same mutation happens in different places at the same time, it's much more likely that the "virus is learning that's a good mutation," explains Kamil.
The process is called convergent evolution, and it was a fascinating topic long before COVID. Just as your heritage can be traced through DNA, so can that of viruses, and when separate lineages develop similar traits it's almost like scientists can see evolution happening in real time. A mutation to SARS-CoV-2 that happens in more than one place at once is a mutation that makes it easier in some way for the virus to survive and that is when it may become alarming. The widespread, documented variants P.1 and B.1.351 are examples of convergence because they share some of the same virulent mutations despite having developed thousands of miles apart.
However, even variants that are emerging in different places at the same time don't present the kind of threat SARS-CoV-2 did in 2019. "This is nature," says Kamil. "It just means that this virus will not easily be driven to extinction or complete elimination by vaccines." Although a person who has already had COVID-19 can be reinfected with a variant, "it is almost always much milder disease" than the original infection, Kamil adds. Rather than causing full-fledged disease, variants have the potiental to "penetrate herd immunity, spreading relatively quietly among people who have developed natural immunity or been vaccinated, until the virus finds someone who has no immunity yet, and that person would be at risk of hospitalization-grade severe disease or death."
Surveillance and predictions
According to Lostroh, genomic surveillance can help scientists predict what's going to happen. "With the British strain, for instance, that's more transmissible, you can measure how fast it's doubling in the population and you can sort of tell whether we should take more measures against this mutation. Should we shut things down a little longer because that mutation is present in the population? That could be really useful if you did enough sampling in the population that you knew where it was," says Lostroh. If, for example, the more transmissible strain was present in 50 percent of cases, but in another county or state it was barely present, it would allow for rolling lockdowns instead of sweeping measures.
Variants are also extremely important when it comes to the development, manufacture, and distribution of vaccines. "You're also looking at medical countermeasures, such as whether your vaccine is still effective, or if your antiviral needs to be updated," says Lane Warmbrod, a senior analyst and research associate at Johns Hopkins Center for Health Security.
Properly funded and extensive genomic surveillance could eventually help control endemic diseases, too, like the seasonal flu, or other common respiratory infections. Kamil says he envisions a future in which genomic surveillance allows for prediction of sickness just as the weather is predicted today. "It's a 51 for infection today at the San Francisco Airport. There's been detection of some respiratory viruses," he says, offering an example. He says that if you're a vulnerable person, if you're immune-suppressed for some reason, you may want to wear a mask based on the sickness report.
The U.S. has the ability, but lacks standards
The benefits of widespread genomic surveillance are clear, and the United States certainly has the necessary technology, equipment, and personnel to carry it out. But, it's not happening at the speed and extent it needs to for the country to gain the benefits.
"The numbers are improving," said Kamil. "We're probably still at less than half a percent of all the samples that have been taken have been sequenced since the beginning of the pandemic."
Although there's no consensus on how many sequences is ideal for a robust surveillance program, modeling performed by the company Illumina suggests about 5 percent of positive tests should be sequenced. The reasons the U.S. has lagged in implementing a sequencing program are complex and varied, but solvable.
Perhaps the most important element that is currently missing is leadership. In order to conduct an effective genomic surveillance program, there need to be standards. The Johns Hopkins Center for Health Security recently published a paper with recommendations as to what kinds of elements need to be standardized in order to make the best use of sequencing technology and analysis.
"Along with which bioinformatic pipelines you're going to use to do the analyses, which sequencing strategy protocol are you going to use, what's your sampling strategy going to be, how is the data is going to be reported, what data gets reported," says Warmbrod. Currently, there's no guidance from the CDC on any of those things. So, while scientists can collect and report information, they may be collecting and reporting different information that isn't comparable, making it less useful for public health measures and vaccine updates.
Globally, one of the most important tools in making the information from genomic surveillance useful is GISAID, a platform designed for scientists to share -- and, importantly, to be credited for -- their data regarding genetic sequences of influenza. Originally, it was launched as a database of bird flu sequences, but has evolved to become an essential tool used by the WHO to make flu vaccine virus recommendations each year. Scientists who share their credentials have free access to the database, and anyone who uses information from the database must credit the scientist who uploaded that information.
Safety, logistics, and funding matter
Scientists at university labs and other small organizations have been uploading sequences to GISAID almost from the beginning of the pandemic, but their funding is generally limited, and there are no standards regarding information collection or reporting. Private, for-profit labs haven't had motivation to set up sequencing programs, although many of them have the logistical capabilities and funding to do so. Public health departments are understaffed, underfunded, and overwhelmed.
University labs may also be limited by safety concerns. The SARS-CoV-2 virus is dangerous, and there's a question of how samples should be transported to labs for sequencing.
Larger, for-profit organizations often have the tools and distribution capabilities to safely collect and sequence samples, but there hasn't been a profit motive. Genomic sequencing is less expensive now than ever before, but even at $100 per sample, the cost adds up -- not to mention the cost of employing a scientist with the proper credentials to analyze the sequence.
The path forward
The recently passed COVID-19 relief bill does have some funding to address genomic sequencing. Specifically, the American Rescue Plan Act includes $1.75 billion in funding for the Centers for Disease Control and Prevention's Advanced Molecular Detection (AMD) program. In an interview last month, CDC Director Rochelle Walensky said that the additional funding will be "a dial. And we're going to need to dial it up." AMD has already announced a collaboration called the Sequencing for Public Health Emergency Response, Epidemiology, and Surveillance (SPHERES) Initiative that will bring together scientists from public health, academic, clinical, and non-profit laboratories across the country with the goal of accelerating sequencing.
Such a collaboration is a step toward following the recommendations in the paper Warmbrod coauthored. Building capacity now, creating a network of labs, and standardizing procedures will mean improved health in the future. "I want to be optimistic," she says. "The good news is there are a lot of passionate, smart, capable people who are continuing to work with government and work with different stakeholders." She cautions, however, that without a national strategy we won't succeed.
"If we maximize the potential and create that framework now, we can also use it for endemic diseases," she says. "It's a very helpful system for more than COVID if we're smart in how we plan it."
Although Jonas Salk has gone down in history for helping rid the world (almost) of polio, his revolutionary vaccine wouldn't have been possible without the world’s largest clinical trial – and the bravery of thousands of kids.
Exactly 67 years ago, in 1955, a group of scientists and reporters gathered at the University of Michigan and waited with bated breath for Dr. Thomas Francis Jr., director of the school’s Poliomyelitis Vaccine Evaluation Center, to approach the podium. The group had gathered to hear the news that seemingly everyone in the country had been anticipating for the past two years – whether the vaccine for poliomyelitis, developed by Francis’s former student Jonas Salk, was effective in preventing the disease.
Polio, at that point, had become a household name. As the highly contagious virus swept through the United States, cities closed their schools, movie theaters, swimming pools, and even churches to stop the spread. For most, polio presented as a mild illness, and was usually completely asymptomatic – but for an unlucky few, the virus took hold of the central nervous system and caused permanent paralysis of muscles in the legs, arms, and even people’s diaphragms, rendering the person unable to walk and breathe. It wasn’t uncommon to hear reports of people – mostly children – who fell sick with a flu-like virus and then, just days later, were relegated to spend the rest of their lives in an iron lung.
For two years, researchers had been testing a vaccine that would hopefully be able to stop the spread of the virus and prevent the 45,000 infections each year that were keeping the nation in a chokehold. At the podium, Francis greeted the crowd and then proceeded to change the course of human history: The vaccine, he reported, was “safe, effective, and potent.” Widespread vaccination could begin in just a few weeks. The nightmare was over.
The road to success
Jonas Salk, a medical researcher and virologist who developed the vaccine with his own research team, would rightfully go down in history as the man who eradicated polio. (Today, wild poliovirus circulates in just two countries, Afghanistan and Pakistan – with only 140 cases reported in 2020.) But many people today forget that the widespread vaccination campaign that effectively ended wild polio across the globe would have never been possible without the human clinical trials that preceded it.
As with the COVID-19 vaccine, skepticism and misinformation around the polio vaccine abounded. But even more pervasive than the skepticism was fear. The consequences of polio had arguably never been more visible.
The road to human clinical trials – and the resulting vaccine – was a long one. In 1938, President Franklin Delano Roosevelt launched the National Foundation for Infantile Paralysis in order to raise funding for research and development of a polio vaccine. (Today, we know this organization as the March of Dimes.) A polio survivor himself, Roosevelt elevated awareness and prevention into the national spotlight, even more so than it had been previously. Raising funds for a safe and effective polio vaccine became a cornerstone of his presidency – and the funds raked in by his foundation went primarily to Salk to fund his research.
The Trials Begin
Salk’s vaccine, which included an inactivated (killed) polio virus, was promising – but now the researchers needed test subjects to make global vaccination a possibility. Because the aim of the vaccine was to prevent paralytic polio, researchers decided that they had to test the vaccine in the population that was most vulnerable to paralysis – young children. And, because the rate of paralysis was so low even among children, the team required many children to collect enough data. Francis, who led the trial to evaluate Salk’s vaccine, began the process of recruiting more than one million school-aged children between the ages of six and nine in 272 counties that had the highest incidence of the disease. The participants were nicknamed the “Polio Pioneers.”
Double-blind, placebo-based trials were considered the “gold standard” of epidemiological research back in Francis's day - and they remain the best approach we have today. These rigorous scientific studies are designed with two participant groups in mind. One group, called the test group, receives the experimental treatment (such as a vaccine); the other group, called the control, receives an inactive treatment known as a placebo. The researchers then compare the effects of the active treatment against the effects of the placebo, and every researcher is “blinded” as to which participants receive what treatment. That way, the results aren’t tainted by any possible biases.
But the study was controversial in that only some of the individual field trials at the county and state levels had a placebo group. Researchers described this as a “calculated risk,” meaning that while there were risks involved in giving the vaccine to a large number of children, the bigger risk was the potential paralysis or death that could come with being infected by polio. In all, just 200,000 children across the US received a placebo treatment, while an additional 725,000 children acted as observational controls – in other words, researchers monitored them for signs of infection, but did not give them any treatment.
As with the COVID-19 vaccine, skepticism and misinformation around the polio vaccine abounded. But even more pervasive than the skepticism was fear. President Roosevelt, who had made many public and televised appearances in a wheelchair, served as a perpetual reminder of the consequences of polio, as an infection at age 39 had rendered him permanently unable to walk. The consequences of polio had arguably never been more visible, and parents signed up their children in droves to participate in the study and offer them protection.
The Polio Pioneer Legacy
In a little less than a year, roughly half a million children received a dose of Salk’s polio vaccine. While plenty of children were hesitant to get the shot, many former participants still remember the fear surrounding the disease. One former participant, a Polio Pioneer named Debbie LaCrosse, writes of her experience: “There was no discussion, no listing of pros and cons. No amount of concern over possible side effects or other unknowns associated with a new vaccine could compare to the terrifying threat of polio.” For their participation, each kid received a certificate – and sometimes a pin – with the words “Polio Pioneer” emblazoned across the front.
When Francis announced the results of the trial on April 12, 1955, people did more than just breathe a sigh of relief – they openly celebrated, ringing church bells and flooding into the streets to embrace. Salk, who had become the face of the vaccine at that point, was instantly hailed as a national hero – and teachers around the country had their students to write him ‘thank you’ notes for his years of diligent work.
But while Salk went on to win national acclaim – even accepting the Presidential Medal of Freedom for his work on the polio vaccine in 1977 – his success was due in no small part to the children (and their parents) who took a risk in order to advance medical science. And that risk paid off: By the early 1960s, the yearly cases of polio in the United States had gone down to just 910. Where before the vaccine polio had caused around 15,000 cases of paralysis each year, only ten cases of paralysis were recorded in the entire country throughout the 1970s. And in 1979, the virus that once shuttered entire towns was declared officially eradicated in this country. Thanks to the efforts of these brave pioneers, the nation – along with the majority of the world – remains free of polio even today.
Why you should (virtually) care
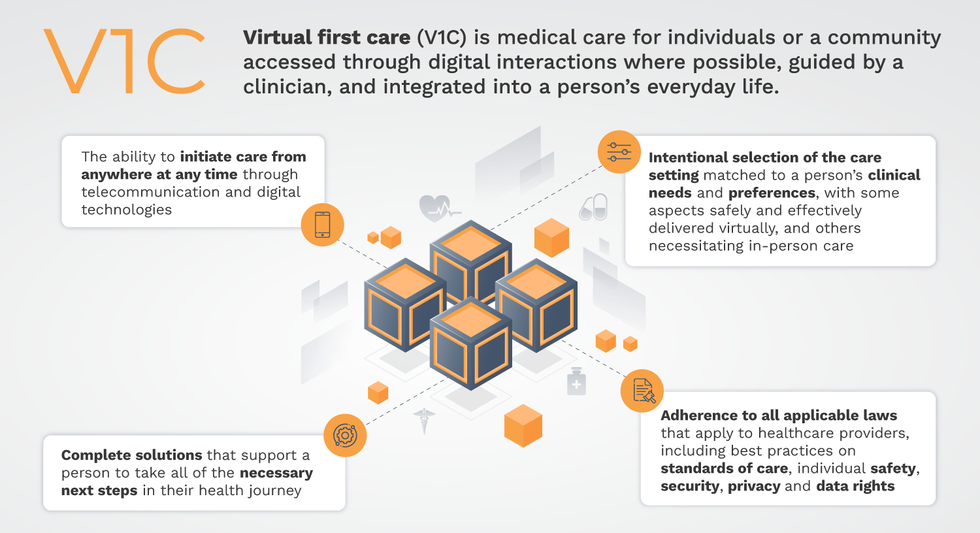
Virtual-first care, or V1C, could increase the quality of healthcare and make it more patient-centric by letting patients combine in-person visits with virtual options such as video for seeing their care providers.
As the pandemic turns endemic, healthcare providers have been eagerly urging patients to return to their offices to enjoy the benefits of in-person care.
But wait.
The last two years have forced all sorts of organizations to be nimble, adaptable and creative in how they work, and this includes healthcare providers’ efforts to maintain continuity of care under the most challenging of conditions. So before we go back to “business as usual,” don’t we owe it to those providers and ourselves to admit that business as usual did not work for most of the people the industry exists to help? If we’re going to embrace yet another period of change – periods that don’t happen often in our complex industry – shouldn’t we first stop and ask ourselves what we’re trying to achieve?
Certainly, COVID has shown that telehealth can be an invaluable tool, particularly for patients in rural and underserved communities that lack access to specialty care. It’s also become clear that many – though not all – healthcare encounters can be effectively conducted from afar. That said, the telehealth tactics that filled the gap during the pandemic were largely stitched together substitutes for existing visit-based workflows: with offices closed, patients scheduled video visits for help managing the side effects of their blood pressure medications or to see their endocrinologist for a quarterly check-in. Anyone whose children slogged through the last year or two of remote learning can tell you that simply virtualizing existing processes doesn’t necessarily improve the experience or the outcomes!
But what if our approach to post-pandemic healthcare came from a patient-driven perspective? We have a fleeting opportunity to advance a care model centered on convenient and equitable access that first prioritizes good outcomes, then selects approaches to care – and locations – tailored to each patient. Using the example of education, imagine how effective it would be if each student, regardless of their school district and aptitude, received such individualized attention.
That’s the idea behind virtual-first care (V1C), a new care model centered on convenient, customized, high-quality care that integrates a full suite of services tailored directly to patients’ clinical needs and preferences. This package includes asynchronous communication such as texting; video and other live virtual modes; and in-person options.
V1C goes beyond what you might think of as standard “telehealth” by using evidence-based protocols and tools that include traditional and digital therapeutics and testing, personalized care plans, dynamic patient monitoring, and team-based approaches to care. This could include spit kits mailed for laboratory tests and complementing clinical care with health coaching. V1C also replaces some in-person exams with ongoing monitoring, using sensors for more ‘whole person’ care.
Amidst all this momentum, we have the opportunity to rethink the goals of healthcare innovation, but that means bringing together key stakeholders to demonstrate that sustainable V1C can redefine healthcare.
Established V1C healthcare providers such as Omada, Headspace, and Heartbeat Health, as well as emerging market entrants like Oshi, Visana, and Wellinks, work with a variety of patients who have complicated long-term conditions such as diabetes, heart failure, gastrointestinal illness, endometriosis, and COPD. V1C is comprehensive in ways that are lacking in digital health and its other predecessors: it has the potential to integrate multiple data streams, incorporate more frequent touches and check-ins over time, and manage a much wider range of chronic health conditions, improving lives and reducing disease burden now and in the future.
Recognizing the pandemic-driven interest in virtual care, significant energy and resources are already flowing fast toward V1C. Some of the world’s largest innovators jumped into V1C early on: Verily, Alphabet’s Life Sciences Company, launched Onduo in 2016 to disrupt the diabetes healthcare market, and is now well positioned to scale its solutions. Major insurers like Aetna and United now offer virtual-first plans to members, responding as organizations expand virtual options for employees. Amidst all this momentum, we have the opportunity to rethink the goals of healthcare innovation, but that means bringing together key stakeholders to demonstrate that sustainable V1C can redefine healthcare.
That was the immediate impetus for IMPACT, a consortium of V1C companies, investors, payers and patients founded last year to ensure access to high-quality, evidence-based V1C. Developed by our team at the Digital Medicine Society (DiMe) in collaboration with the American Telemedicine Association (ATA), IMPACT has begun to explore key issues that include giving patients more integrated experiences when accessing both virtual and brick-and-mortar care.
Digital Medicine Society
V1C is not, nor should it be, virtual-only care. In this new era of hybrid healthcare, success will be defined by how well providers help patients navigate the transitions. How do we smoothly hand a patient off from an onsite primary care physician to, say, a virtual cardiologist? How do we get information from a brick-and-mortar to a digital portal? How do you manage dataflow while still staying HIPAA compliant? There are many complex regulatory implications for these new models, as well as an evolving landscape in terms of privacy, security and interoperability. It will be no small task for groups like IMPACT to determine the best path forward.
None of these factors matter unless the industry can recruit and retain clinicians. Our field is facing an unprecedented workforce crisis. Traditional healthcare is making clinicians miserable, and COVID has only accelerated the trend of overworked, disenchanted healthcare workers leaving in droves. Clinicians want more interactions with patients, and fewer with computer screens – call it “More face time, less FaceTime.” No new model will succeed unless the industry can more efficiently deploy its talent – arguably its most scarce and precious resource. V1C can help with alleviating the increasing burden and frustration borne by individual physicians in today’s status quo.
In healthcare, new technological approaches inevitably provoke no shortage of skepticism. Past lessons from Silicon Valley-driven fixes have led to understandable cynicism. But V1C is a different breed of animal. By building healthcare around the patient, not the clinic, V1C can make healthcare work better for patients, payers and providers. We’re at a fork in the road: we can revert back to a broken sick-care system, or dig in and do the hard work of figuring out how this future-forward healthcare system gets financed, organized and executed. As a field, we must find the courage and summon the energy to embrace this moment, and make it a moment of change.

