COVID Variants Are Like “a Thief Changing Clothes” – and Our Camera System Barely Exists

Being able to track variants of concern in real time is crucial to our ability to stay ahead of the virus.
Whether it's "natural selection" as Darwin called it, or it's "mutating" as the X-Men called it, living organisms change over time, developing thumbs or more efficient protein spikes, depending on the organism and the demands of its environment. The coronavirus that causes COVID-19, SARS-CoV-2, is not an exception, and now, after the virus has infected millions of people around the globe for more than a year, scientists are beginning to see those changes.
The notorious variants that have popped up include B.1.1.7, sometimes called the UK variant, as well as P.1 and B.1.351, which seem to have emerged in Brazil and South Africa respectively. As vaccinations are picking up pace, officials are warning that now
is not the time to become complacent or relax restrictions because the variants aren't well understood.
Some appear to be more transmissible, and deadlier, while others can evade the immune system's defenses better than earlier versions of the virus, potentially undermining the effectiveness of vaccines to some degree. Genomic surveillance, the process of sequencing the genetic code of the virus widely to observe changes and patterns, is a critical way that scientists can keep track of its evolution and work to understand how the variants might affect humans.
"It's like a thief changing clothes"
It's important to note that viruses mutate all the time. If there were funding and personnel to sequence the genome of every sample of the virus, scientists would see thousands of mutations. Not every variant deserves our attention. The vast majority of mutations are not important at all, but recognizing those that are is a crucial tool in getting and staying ahead of the virus. The work of sequencing, analyzing, observing patterns, and using public health tools as necessary is complicated and confusing to those without years of specialized training.
Jeremy Kamil, associate professor of microbiology and immunology at LSU Health Shreveport, in Louisiana, says that the variants developing are like a thief changing clothes. The thief goes in your house, steals your stuff, then leaves and puts on a different shirt and a wig, in the hopes you won't recognize them. Genomic surveillance catches the "thief" even in those different clothes.
One of the tricky things about variants is recognizing the point at which they move from interesting, to concerning at a local level, to dangerous in a larger context.
Understanding variants, both the uninteresting ones and the potentially concerning ones, gives public health officials and researchers at different levels a useful set of tools. Locally, knowing which variants are circulating in the community helps leaders know whether mask mandates and similar measures should be implemented or discontinued, or whether businesses and schools can open relatively safely.
There's more to it than observing new variants
Analysis is complex, particularly when it comes to understanding which variants are of concern. "So the question is always if a mutation becomes common, is that a random occurrence?" says Phoebe Lostroh, associate professor of molecular biology at Colorado College. "Or is the variant the result of some kind of selection because the mutation changes some property about the virus that makes it reproduce more quickly than variants of the virus that don't have that mutation? For a virus, [mutations can affect outcomes like] how much it replicates inside a person's body, how much somebody breathes it out, whether the particles that somebody might breathe in get smaller and can lead to greater transmission."
Along with all of those factors, accurate and useful genomic surveillance requires an understanding of where variants are occurring, how they are related, and an examination of why they might be prevalent.
For example, if a potentially worrisome variant appears in a community and begins to spread very quickly, it's not time to raise a public health alarm until several important questions have been answered, such as whether the variant is spreading due to specific events, or if it's happening because the mutation has allowed the virus to infect people more efficiently. Kamil offered a hypothetical scenario to explain: Imagine that a member of a community became infected and the virus mutated. That person went to church and three more people were infected, but one of them went to a karaoke bar and while singing infected 100 other people. Examining the conditions under which the virus has spread is, therefore, an essential part of untangling whether a mutation itself made the virus more transmissible or if an infected person's behaviors contributed to a local outbreak.
One of the tricky things about variants is recognizing the point at which they move from interesting, to concerning at a local level, to dangerous in a larger context. Genomic sequencing can help with that, but only when it's coordinated. When the same mutation occurs frequently, but is localized to one region, it's a concern, but when the same mutation happens in different places at the same time, it's much more likely that the "virus is learning that's a good mutation," explains Kamil.
The process is called convergent evolution, and it was a fascinating topic long before COVID. Just as your heritage can be traced through DNA, so can that of viruses, and when separate lineages develop similar traits it's almost like scientists can see evolution happening in real time. A mutation to SARS-CoV-2 that happens in more than one place at once is a mutation that makes it easier in some way for the virus to survive and that is when it may become alarming. The widespread, documented variants P.1 and B.1.351 are examples of convergence because they share some of the same virulent mutations despite having developed thousands of miles apart.
However, even variants that are emerging in different places at the same time don't present the kind of threat SARS-CoV-2 did in 2019. "This is nature," says Kamil. "It just means that this virus will not easily be driven to extinction or complete elimination by vaccines." Although a person who has already had COVID-19 can be reinfected with a variant, "it is almost always much milder disease" than the original infection, Kamil adds. Rather than causing full-fledged disease, variants have the potiental to "penetrate herd immunity, spreading relatively quietly among people who have developed natural immunity or been vaccinated, until the virus finds someone who has no immunity yet, and that person would be at risk of hospitalization-grade severe disease or death."
Surveillance and predictions
According to Lostroh, genomic surveillance can help scientists predict what's going to happen. "With the British strain, for instance, that's more transmissible, you can measure how fast it's doubling in the population and you can sort of tell whether we should take more measures against this mutation. Should we shut things down a little longer because that mutation is present in the population? That could be really useful if you did enough sampling in the population that you knew where it was," says Lostroh. If, for example, the more transmissible strain was present in 50 percent of cases, but in another county or state it was barely present, it would allow for rolling lockdowns instead of sweeping measures.
Variants are also extremely important when it comes to the development, manufacture, and distribution of vaccines. "You're also looking at medical countermeasures, such as whether your vaccine is still effective, or if your antiviral needs to be updated," says Lane Warmbrod, a senior analyst and research associate at Johns Hopkins Center for Health Security.
Properly funded and extensive genomic surveillance could eventually help control endemic diseases, too, like the seasonal flu, or other common respiratory infections. Kamil says he envisions a future in which genomic surveillance allows for prediction of sickness just as the weather is predicted today. "It's a 51 for infection today at the San Francisco Airport. There's been detection of some respiratory viruses," he says, offering an example. He says that if you're a vulnerable person, if you're immune-suppressed for some reason, you may want to wear a mask based on the sickness report.
The U.S. has the ability, but lacks standards
The benefits of widespread genomic surveillance are clear, and the United States certainly has the necessary technology, equipment, and personnel to carry it out. But, it's not happening at the speed and extent it needs to for the country to gain the benefits.
"The numbers are improving," said Kamil. "We're probably still at less than half a percent of all the samples that have been taken have been sequenced since the beginning of the pandemic."
Although there's no consensus on how many sequences is ideal for a robust surveillance program, modeling performed by the company Illumina suggests about 5 percent of positive tests should be sequenced. The reasons the U.S. has lagged in implementing a sequencing program are complex and varied, but solvable.
Perhaps the most important element that is currently missing is leadership. In order to conduct an effective genomic surveillance program, there need to be standards. The Johns Hopkins Center for Health Security recently published a paper with recommendations as to what kinds of elements need to be standardized in order to make the best use of sequencing technology and analysis.
"Along with which bioinformatic pipelines you're going to use to do the analyses, which sequencing strategy protocol are you going to use, what's your sampling strategy going to be, how is the data is going to be reported, what data gets reported," says Warmbrod. Currently, there's no guidance from the CDC on any of those things. So, while scientists can collect and report information, they may be collecting and reporting different information that isn't comparable, making it less useful for public health measures and vaccine updates.
Globally, one of the most important tools in making the information from genomic surveillance useful is GISAID, a platform designed for scientists to share -- and, importantly, to be credited for -- their data regarding genetic sequences of influenza. Originally, it was launched as a database of bird flu sequences, but has evolved to become an essential tool used by the WHO to make flu vaccine virus recommendations each year. Scientists who share their credentials have free access to the database, and anyone who uses information from the database must credit the scientist who uploaded that information.
Safety, logistics, and funding matter
Scientists at university labs and other small organizations have been uploading sequences to GISAID almost from the beginning of the pandemic, but their funding is generally limited, and there are no standards regarding information collection or reporting. Private, for-profit labs haven't had motivation to set up sequencing programs, although many of them have the logistical capabilities and funding to do so. Public health departments are understaffed, underfunded, and overwhelmed.
University labs may also be limited by safety concerns. The SARS-CoV-2 virus is dangerous, and there's a question of how samples should be transported to labs for sequencing.
Larger, for-profit organizations often have the tools and distribution capabilities to safely collect and sequence samples, but there hasn't been a profit motive. Genomic sequencing is less expensive now than ever before, but even at $100 per sample, the cost adds up -- not to mention the cost of employing a scientist with the proper credentials to analyze the sequence.
The path forward
The recently passed COVID-19 relief bill does have some funding to address genomic sequencing. Specifically, the American Rescue Plan Act includes $1.75 billion in funding for the Centers for Disease Control and Prevention's Advanced Molecular Detection (AMD) program. In an interview last month, CDC Director Rochelle Walensky said that the additional funding will be "a dial. And we're going to need to dial it up." AMD has already announced a collaboration called the Sequencing for Public Health Emergency Response, Epidemiology, and Surveillance (SPHERES) Initiative that will bring together scientists from public health, academic, clinical, and non-profit laboratories across the country with the goal of accelerating sequencing.
Such a collaboration is a step toward following the recommendations in the paper Warmbrod coauthored. Building capacity now, creating a network of labs, and standardizing procedures will mean improved health in the future. "I want to be optimistic," she says. "The good news is there are a lot of passionate, smart, capable people who are continuing to work with government and work with different stakeholders." She cautions, however, that without a national strategy we won't succeed.
"If we maximize the potential and create that framework now, we can also use it for endemic diseases," she says. "It's a very helpful system for more than COVID if we're smart in how we plan it."
A company in England has made a test that picks out the compounds from breath that reveal if people have liver disease.
Every year, around two million people worldwide die of liver disease. While some people inherit the disease, it’s most commonly caused by hepatitis, obesity and alcoholism. These underlying conditions kill liver cells, causing scar tissue to form until eventually the liver cannot function properly. Since 1979, deaths due to liver disease have increased by 400 percent.
The sooner the disease is detected, the more effective treatment can be. But once symptoms appear, the liver is already damaged. Around 50 percent of cases are diagnosed only after the disease has reached the final stages, when treatment is largely ineffective.
To address this problem, Owlstone Medical, a biotech company in England, has developed a breath test that can detect liver disease earlier than conventional approaches. Human breath contains volatile organic compounds (VOCs) that change in the first stages of liver disease. Owlstone’s breath test can reliably collect, store and detect VOCs, while picking out the specific compounds that reveal liver disease.
“There’s a need to screen more broadly for people with early-stage liver disease,” says Owlstone’s CEO Billy Boyle. “Equally important is having a test that's non-invasive, cost effective and can be deployed in a primary care setting.”
The standard tool for detection is a biopsy. It is invasive and expensive, making it impractical to use for people who aren't yet symptomatic. Meanwhile, blood tests are less invasive, but they can be inaccurate and can’t discriminate between different stages of the disease.
In the past, breath tests have not been widely used because of the difficulties of reliably collecting and storing breath. But Owlstone’s technology could help change that.
The team is testing patients in the early stages of advanced liver disease, or cirrhosis, to identify and detect these biomarkers. In an initial study, Owlstone’s breathalyzer was able to pick out patients who had early cirrhosis with 83 percent sensitivity.
Boyle’s work is personally motivated. His wife died of colorectal cancer after she was diagnosed with a progressed form of the disease. “That was a big impetus for me to see if this technology could work in early detection,” he says. “As a company, Owlstone is interested in early detection across a range of diseases because we think that's a way to save lives and a way to save costs.”
How it works
In the past, breath tests have not been widely used because of the difficulties of reliably collecting and storing breath. But Owlstone’s technology could help change that.
Study participants breathe into a mouthpiece attached to a breath sampler developed by Owlstone. It has cartridges are designed and optimized to collect gases. The sampler specifically targets VOCs, extracting them from atmospheric gases in breath, to ensure that even low levels of these compounds are captured.
The sampler can store compounds stably before they are assessed through a method called mass spectrometry, in which compounds are converted into charged atoms, before electromagnetic fields filter and identify even the tiniest amounts of charged atoms according to their weight and charge.
The top four compounds in our breath
In an initial study, Owlstone captured VOCs in breath to see which ones could help them tell the difference between people with and without liver disease. They tested the breath of 46 patients with liver disease - most of them in the earlier stages of cirrhosis - and 42 healthy people. Using this data, they were able to create a diagnostic model. Individually, compounds like 2-Pentanone and limonene performed well as markers for liver disease. Owlstone achieved even better performance by examining the levels of the top four compounds together, distinguishing between liver disease cases and controls with 95 percent accuracy.
“It was a good proof of principle since it looks like there are breath biomarkers that can discriminate between diseases,” Boyle says. “That was a bit of a stepping stone for us to say, taking those identified, let’s try and dose with specific concentrations of probes. It's part of building the evidence and steering the clinical trials to get to liver disease sensitivity.”
Sabine Szunerits, a professor of chemistry in Institute of Electronics at the University of Lille, sees the potential of Owlstone’s technology.
“Breath analysis is showing real promise as a clinical diagnostic tool,” says Szunerits, who has no ties with the company. “Owlstone Medical’s technology is extremely effective in collecting small volatile organic biomarkers in the breath. In combination with pattern recognition it can give an answer on liver disease severity. I see it as a very promising way to give patients novel chances to be cured.”
Improving the breath sampling process
Challenges remain. With more than one thousand VOCs found in the breath, it can be difficult to identify markers for liver disease that are consistent across many patients.
Julian Gardner is a professor of electrical engineering at Warwick University who researches electronic sensing devices. “Everyone’s breath has different levels of VOCs and different ones according to gender, diet, age etc,” Gardner says. “It is indeed very challenging to selectively detect the biomarkers in the breath for liver disease.”
So Owlstone is putting chemicals in the body that they know interact differently with patients with liver disease, and then using the breath sampler to measure these specific VOCs. The chemicals they administer are called Exogenous Volatile Organic Compound) probes, or EVOCs.
Most recently, they used limonene as an EVOC probe, testing 29 patients with early cirrhosis and 29 controls. They gave the limonene to subjects at specific doses to measure how its concentrations change in breath. The aim was to try and see what was happening in their livers.
“They are proposing to use drugs to enhance the signal as they are concerned about the sensitivity and selectivity of their method,” Gardner says. “The approach of EVOC probes is probably necessary as you can then eliminate the person-to-person variation that will be considerable in the soup of VOCs in our breath.”
Through these probes, Owlstone could identify patients with liver disease with 83 percent sensitivity. By targeting what they knew was a disease mechanism, they were able to amplify the signal. The company is starting a larger clinical trial, and the plan is to eventually use a panel of EVOC probes to make sure they can see diverging VOCs more clearly.
“I think the approach of using probes to amplify the VOC signal will ultimately increase the specificity of any VOC breath tests, and improve their practical usability,” says Roger Yazbek, who leads the South Australian Breath Analysis Research (SABAR) laboratory in Flinders University. “Whilst the findings are interesting, it still is only a small cohort of patients in one location.”
The future of breath diagnosis
Owlstone wants to partner with pharmaceutical companies looking to learn if their drugs have an effect on liver disease. They’ve also developed a microchip, a miniaturized version of mass spectrometry instruments, that can be used with the breathalyzer. It is less sensitive but will enable faster detection.
Boyle says the company's mission is for their tests to save 100,000 lives. "There are lots of risks and lots of challenges. I think there's an opportunity to really establish breath as a new diagnostic class.”
Doctors worry that fungal pathogens may cause the next pandemic.
Bacterial antibiotic resistance has been a concern in the medical field for several years. Now a new, similar threat is arising: drug-resistant fungal infections. The Centers for Disease Control and Prevention considers antifungal and antimicrobial resistance to be among the world’s greatest public health challenges.
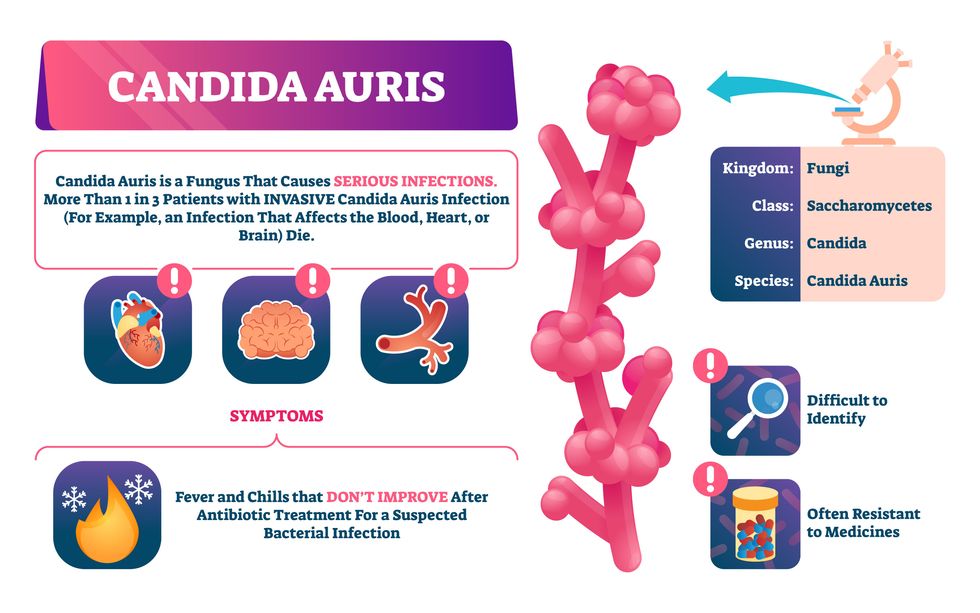
One particular type of fungal infection caused by Candida auris is escalating rapidly throughout the world. And to make matters worse, C. auris is becoming increasingly resistant to current antifungal medications, which means that if you develop a C. auris infection, the drugs your doctor prescribes may not work. “We’re effectively out of medicines,” says Thomas Walsh, founding director of the Center for Innovative Therapeutics and Diagnostics, a translational research center dedicated to solving the antimicrobial resistance problem. Walsh spoke about the challenges at a Demy-Colton Virtual Salon, one in a series of interactive discussions among life science thought leaders.
Although C. auris typically doesn’t sicken healthy people, it afflicts immunocompromised hospital patients and may cause severe infections that can lead to sepsis, a life-threatening condition in which the overwhelmed immune system begins to attack the body’s own organs. Between 30 and 60 percent of patients who contract a C. auris infection die from it, according to the CDC. People who are undergoing stem cell transplants, have catheters or have taken antifungal or antibiotic medicines are at highest risk. “We’re coming to a perfect storm of increasing resistance rates, increasing numbers of immunosuppressed patients worldwide and a bug that is adapting to higher temperatures as the climate changes,” says Prabhavathi Fernandes, chair of the National BioDefense Science Board.
Most Candida species aren’t well-adapted to our body temperatures so they aren’t a threat. C. auris, however, thrives at human body temperatures.
Although medical professionals aren’t concerned at this point about C. auris evolving to affect healthy people, they worry that its presence in hospitals can turn routine surgeries into life-threatening calamities. “It’s coming,” says Fernandes. “It’s just a matter of time.”
An emerging global threat
“Fungi are found in the environment,” explains Fernandes, so Candida spores can easily wind up on people’s skin. In hospitals, they can be transferred from contact with healthcare workers or contaminated surfaces. Most Candida species aren’t well-adapted to our body temperatures so they aren’t a threat. C. auris, however, thrives at human body temperatures. It can enter the body during medical treatments that break the skin—and cause an infection. Overall, fungal infections cost some $48 billion in the U.S. each year. And infection rates are increasing because, in an ironic twist, advanced medical therapies are enabling severely ill patients to live longer and, therefore, be exposed to this pathogen.
The first-ever case of a C. auris infection was reported in Japan in 2009, although an analysis of Candida samples dated the earliest strain to a 1996 sample from South Korea. Since then, five separate varieties – called clades, which are similar to strains among bacteria – developed independently in different geographies: South Asia, East Asia, South Africa, South America and, recently, Iran. So far, C. auris infections have been reported in 35 countries.
In the U.S., the first infection was reported in 2016, and the CDC started tracking it nationally two years later. During that time, 5,654 cases have been reported to the CDC, which only tracks U.S. data.
What’s more notable than the number of cases is their rate of increase. In 2016, new cases increased by 175 percent and, on average, they have approximately doubled every year. From 2016 through 2022, the number of infections jumped from 63 to 2,377, a roughly 37-fold increase.
“This reminds me of what we saw with epidemics from 2013 through 2020… with Ebola, Zika and the COVID-19 pandemic,” says Robin Robinson, CEO of Spriovas and founding director of the Biomedical Advanced Research and Development Authority (BARDA), which is part of the U.S. Department of Health and Human Services. These epidemics started with a hockey stick trajectory, Robinson says—a gradual growth leading to a sharp spike, just like the shape of a hockey stick.
Another challenge is that right now medics don’t have rapid diagnostic tests for fungal infections. Currently, patients are often misdiagnosed because C. auris resembles several other easily treated fungi. Or they are diagnosed long after the infection begins and is harder to treat.
The problem is that existing diagnostics tests can only identify C. auris once it reaches the bloodstream. Yet, because this pathogen infects bodily tissues first, it should be possible to catch it much earlier before it becomes life-threatening. “We have to diagnose it before it reaches the bloodstream,” Walsh says.
The most alarming fact is that some Candida infections no longer respond to standard therapeutics.
“We need to focus on rapid diagnostic tests that do not rely on a positive blood culture,” says John Sperzel, president and CEO of T2 Biosystems, a company specializing in diagnostics solutions. Blood cultures typically take two to three days for the concentration of Candida to become large enough to detect. The company’s novel test detects about 90 percent of Candida species within three to five hours—thanks to its ability to spot minute quantities of the pathogen in blood samples instead of waiting for them to incubate and proliferate.
Unlike other Candida species C. auris thrives at human body temperatures
Adobe Stock
Tackling the resistance challenge
The most alarming fact is that some Candida infections no longer respond to standard therapeutics. The number of cases that stopped responding to echinocandin, the first-line therapy for most Candida infections, tripled in 2020, according to a study by the CDC.
Now, each of the first four clades shows varying levels of resistance to all three commonly prescribed classes of antifungal medications, such as azoles, echinocandins, and polyenes. For example, 97 percent of infections from C. auris Clade I are resistant to fluconazole, 54 percent to voriconazole and 30 percent of amphotericin. Nearly half are resistant to multiple antifungal drugs. Even with Clade II fungi, which has the least resistance of all the clades, 11 to 14 percent have become resistant to fluconazole.
Anti-fungal therapies typically target specific chemical compounds present on fungi’s cell membranes, but not on human cells—otherwise the medicine would cause damage to our own tissues. Fluconazole and other azole antifungals target a compound called ergosterol, preventing the fungal cells from replicating. Over the years, however, C. auris evolved to resist it, so existing fungal medications don’t work as well anymore.
A newer class of drugs called echinocandins targets a different part of the fungal cell. “The echinocandins – like caspofungin – inhibit (a part of the fungi) involved in making glucan, which is an essential component of the fungal cell wall and is not found in human cells,” Fernandes says. New antifungal treatments are needed, she adds, but there are only a few magic bullets that will hit just the fungus and not the human cells.
Research to fight infections also has been challenged by a lack of government support. That is changing now that BARDA is requesting proposals to develop novel antifungals. “The scope includes C. auris, as well as antifungals following a radiological/nuclear emergency, says BARDA spokesperson Elleen Kane.
The remaining challenge is the number of patients available to participate in clinical trials. Large numbers are needed, but the available patients are quite sick and often die before trials can be completed. Consequently, few biopharmaceutical companies are developing new treatments for C. auris.
ClinicalTrials.gov reports only two drugs in development for invasive C. auris infections—those than can spread throughout the body rather than localize in one particular area, like throat or vaginal infections: ibrexafungerp by Scynexis, Inc., fosmanogepix, by Pfizer.
Scynexis’ ibrexafungerp appears active against C. auris and other emerging, drug-resistant pathogens. The FDA recently approved it as a therapy for vaginal yeast infections and it is undergoing Phase III clinical trials against invasive candidiasis in an attempt to keep the infection from spreading.
“Ibreafungerp is structurally different from other echinocandins,” Fernandes says, because it targets a different part of the fungus. “We’re lucky it has activity against C. auris.”
Pfizer’s fosmanogepix is in Phase II clinical trials for patients with invasive fungal infections caused by multiple Candida species. Results are showing significantly better survival rates for people taking fosmanogepix.
Although C. auris does pose a serious threat to healthcare worldwide, scientists try to stay optimistic—because they recognized the problem early enough, they might have solutions in place before the perfect storm hits. “There is a bit of hope,” says Robinson. “BARDA has finally been able to fund the development of new antifungal agents and, hopefully, this year we can get several new classes of antifungals into development.”

