An At-Home Contagiousness Test for COVID-19 Already Exists. Why Can’t We Use It?
Kira Peikoff was the editor-in-chief of Leaps.org from 2017 to 2021. As a journalist, her work has appeared in The New York Times, Newsweek, Nautilus, Popular Mechanics, The New York Academy of Sciences, and other outlets. She is also the author of four suspense novels that explore controversial issues arising from scientific innovation: Living Proof, No Time to Die, Die Again Tomorrow, and Mother Knows Best. Peikoff holds a B.A. in Journalism from New York University and an M.S. in Bioethics from Columbia University. She lives in New Jersey with her husband and two young sons. Follow her on Twitter @KiraPeikoff.

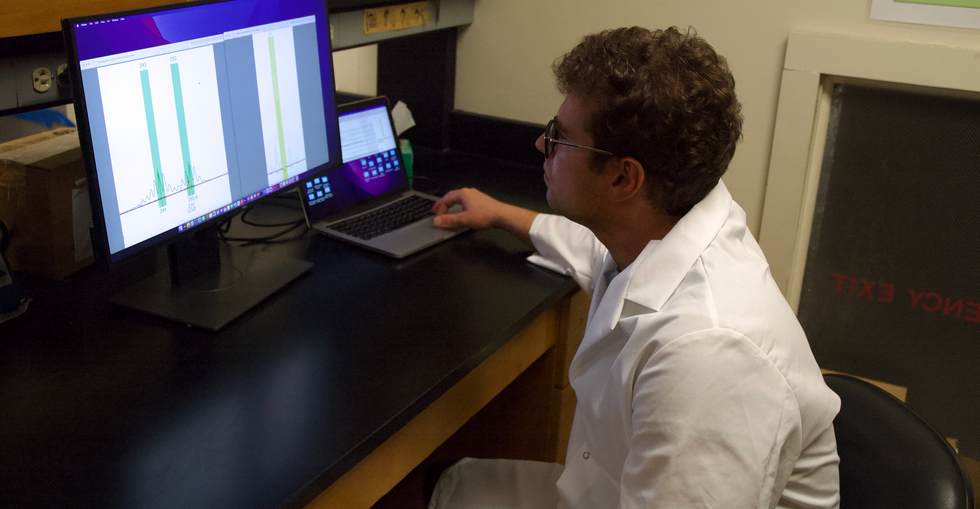
Bobby Brooke Herrera, the co-founder and CEO of e25Bio, demonstrates the company's rapid paper-strip test for detecting the coronavirus.
You're lying in bed late at night, the foggy swirl of the pandemic's 8th month just beginning to fall behind you, when you detect a slight tickle at the back of your throat.
"If half of people choose to use these tests every other day, then we can stop transmission faster than a vaccine can."
Suddenly fully awake, a jolt of panicked electricity races through your body. Has COVID-19 come for you? In the U.S., answering this simple question is incredibly difficult.
Now, you might have to wait for hours in line in your car to get a test for $100, only to find out your result 10-14 days later -- much too late to matter in stopping an outbreak. Due to such obstacles, a recent report in JAMA Internal Medicine estimated that 9 out of 10 infections in the U.S. are being missed.
But what if you could use a paper strip in the privacy of your own home, like a pregnancy test, and find out if you are contagious in real time?
e25 Bio, a small company in Cambridge, Mass., has already created such a test and it has been sitting on a lab bench, inaccessible, since April. It is an antigen test, which looks for proteins on the outside of a virus, and can deliver results in about 15 minutes. Also like an over-the-counter pregnancy test, e25 envisions its paper strips as a public health screening tool, rather than a definitive diagnostic test. People who see a positive result would be encouraged to then seek out a physician-administered, gold-standard diagnostic test: the more sensitive PCR.
Typically, hospitals and other health facilities rely on PCR tests to diagnose viruses. This test can detect small traces of genetic material that a virus leaves behind in the human body, which tells a clinician that the patient is either actively infected with or recently cleared that virus. PCR is quite sensitive, meaning that it is able to detect the presence of a virus' genetic material very accurately.
But although PCR is the gold-standard for diagnostics, it's also the most labor-intensive way to test for a virus and takes a relatively long time to produce results. That's not a good match for stopping super-spreader events during an unchecked pandemic. PCR is also not great at identifying the infected people when they are most at risk of potentially transmitting the virus to others.
That's because the viral threshold at which PCR can detect a positive result is so low, that it's actually too sensitive for the purposes of telling whether someone is contagious.
"The majority of time someone is PCR positive, those [genetic] remnants do not indicate transmissible virus," epidemiologist Michael Mina recently Tweeted. "They indicate remnants of a recently cleared infection."
To stop the chain of transmission for COVID-19, he says, "We need a more accurate test than PCR, that turns positive when someone is able to transmit."
In other words, we need a test that is better at detecting whether a person is contagious, as opposed to whether a small amount of virus can be detected in their nose or saliva. This kind of test is especially critical given the research showing that asymptomatic and pre-symptomatic people have high viral loads and are spreading the virus undetected.
The critical question for contagiousness testing, then, is how big a dose of SARS-CoV-2, the virus that causes COVID, does it take to infect most people? Researchers are still actively trying to answer this. As Angela Rasmussen, a coronavirus expert at Columbia University, told STAT: "We don't know the amount that is required to cause an infection, but it seems that it's probably not a really, really small amount, like measles."
Amesh Adalja, an infectious disease physician and a senior scholar at the Johns Hopkins University Center for Health Security, told LeapsMag: "It's still unclear what viral load is associated with contagiousness but it is biologically plausible that higher viral loads, in general, are associated with more efficient transmission especially in symptomatic individuals. In those without symptoms, however, the same relationship may not hold and this may be one of the reasons young children, despite their high viral loads, are not driving outbreaks."
"Antigen tests work best when there's high viral loads. They're catching people who are super spreaders."
Mina and colleagues estimate that widespread use of weekly cheap, rapid tests that are 100 times less sensitive than PCR tests would prevent outbreaks -- as long as the people who are positive self-isolate.
So why can't we buy e25Bio's test at a drugstore right now? Ironically, it's barred for the very reason that it's useful in the first place: Because it is not sensitive enough to satisfy the U.S. Food and Drug Administration, according to the company.
"We're ready to go," says Carlos-Henri Ferré, senior associate of operations and communications at e25. "We've applied to FDA, and now it's in their hands."
The problem, he said, is that the FDA is evaluating applications for antigen tests based on criteria for assessing diagnostics, like PCR, even when the tests serve a different purpose -- as a screening tool.
"Antigen tests work best when there's high viral loads," Ferré says. "They're catching people who are super spreaders, that are capable of continuing the spread of disease … FDA criteria is for diagnostics and not this."
FDA released guidance on July 29th -- 140 days into the pandemic -- recommending that at-home tests should perform with at least 80 percent sensitivity if ordered by prescription, and at least 90 percent sensitivity if purchased over the counter. "The danger of a false negative result is that it can contribute to the spread of COVID-19," according to an FDA spokesperson. "However, oversight of a health care professional who reviews the results, in combination with the patient's symptoms and uses their clinical judgment to recommend additional testing, if needed, among other things, can help mitigate some risks."
Crucially, the 90 percent sensitivity recommendation is judged upon comparison to PCR tests, meaning that if a PCR test is able to detect virus in 100 samples, the at-home antigen test would need to detect virus in at least 90 of those samples. Since antigen tests only detect high viral loads, frustrated critics like Mina say that such guidance is "unreasonable."
"The FDA at this moment is not understanding the true potential for wide-scale frequent testing. In some ways this is not their fault," Mina told LeapsMag. "The FDA does not have any remit to evaluate tests that fall outside of medical diagnostic testing. The proposal I have put forth is not about diagnostic testing (leave that for symptomatic cases reporting to their physician and getting PCR tests)....Daily rapid tests are not about diagnosing people and they are not about public health surveillance and they are not about passports to go to school, out to dinner or into the office. They are about reducing population-level transmission given a similar approach as vaccines."
A reasonable standard, he added, would be to follow the World Health Organization's Target Product Profiles, which are documents to help developers build desirable and minimally acceptable testing products. "A decent limit," Mina says, "is a 70% or 80% sensitivity (if they truly require sensitivity as a metric) to detect virus at Ct values less than 25. This coincides with detection of the most transmissible people, which is important."
(A Ct value is a type of measurement that corresponds inversely to the amount of viral load in a given sample. Researchers have found that Ct values of 13-17 indicate high viral load, whereas Ct values greater than 34 indicate a lack of infectious virus.)
"We believe this should be an at-home test, but [if FDA approval comes through] the first rollout is to do this in laboratories, hospitals, and clinics."
"We believe that population screening devices have an immediate place and use in helping beat the virus," says Ferré. "You can have a significant impact even with a test at 60% sensitivity if you are testing frequently."
When presented with criticism of its recommendations, the FDA indicated that it will not automatically deny any at-home test that fails to meet the 90 percent sensitivity guidance.
"FDA is always open to alternative proposals from developers, including strategies for serial testing with less sensitive tests," a spokesperson wrote in a statement. "For example, it is possible that overall sensitivity of the strategy could be considered cumulatively rather than based on one-time testing….In the case of a manufacturer with an at-home test that can only detect people with COVID-19 when they have a high viral load, we encourage them to talk with us so we can better understand their test, how they propose to use it, and the validation data they have collected to support that use."
However, the FDA's actions so far conflict with its stated openness. e25 ended up adding a step to the protocol in order to better meet FDA standards for sensitivity, but that extra step—sending samples to a laboratory for results—will undercut the test's ability to work as an at-home screening tool.
"We believe this should be an at-home test, but [if FDA approval comes through] the first rollout is to do this in laboratories, hospitals, and clinics," Ferré says.
According to the FDA, no test developers have approached them with a request for an emergency use authorization that proposes an alternate testing paradigm, such as serial testing, to mitigate test sensitivity below 80 percent.
From a scientific perspective, antigen tests like e25Bio's are not the only horse in the race for a simple rapid test with potential for at-home use. CRISPR technology has long been touted as fertile ground for diagnostics, and in an eerily prescient interview with LeapsMag in November, CRISPR pioneer Feng Zhang spoke of its potential application as an at-home diagnostic for an infectious disease specifically.
"I think in the long run it will be great to see this for, say, at-home disease testing, for influenza and other sorts of important public health [concerns]," he said in the fall. "To be able to get a readout at home, people can potentially quarantine themselves rather than traveling to a hospital and then carrying the risk of spreading that disease to other people as they get to the clinic."
Zhang's company Sherlock Biosciences is now working on scaled-up manufacturing of a test to detect SARS CoV-2. Mammoth Biosciences, which secured funding from the National Institutes of Health's Rapid Acceleration of Diagnostics program, is also working on a CRISPR diagnostic for SARS CoV-2. Both would check the box for rapid testing, but so far not for at-home testing, as they would also require laboratory infrastructure to provide results.
If any at-home tests can clear the regulatory hurdles, they would also need to be manufactured on a large scale and be cheap enough to entice people to actually use them. In the world of at-home diagnostics, pregnancy tests have become the sole mainstream victor because they're simple to use, small to carry, easy to interpret, and costs about seven or eight dollars at any ubiquitous store, like Target or Walmart. By comparison, the at-home COVID collection tests that don't even offer diagnostics—you send away your sample to an external lab—all cost over $100 to take just one time.
For the time being, the only available diagnostics for COVID require a lab or an expensive dedicated machine to process. This disconnect could prolong the world's worst health crisis in a century.
"Daily rapid tests have enormous potential to sever transmission chains and create herd effects similar to herd immunity," Mina says. "We all recognize that vaccines and infections can result in herd immunity when something around half of people are no longer susceptible.
"The same thing exists with these tests. These are the intervention to stop the virus. If half of people choose to use these tests every other day, then we can stop transmission faster than a vaccine can. The technology exists, the theory and mathematics back it up, the epidemiology is sound. There is no reason we are not approaching this as strongly as we would be approaching vaccines."
--Additional reporting by Julia Sklar
Kira Peikoff was the editor-in-chief of Leaps.org from 2017 to 2021. As a journalist, her work has appeared in The New York Times, Newsweek, Nautilus, Popular Mechanics, The New York Academy of Sciences, and other outlets. She is also the author of four suspense novels that explore controversial issues arising from scientific innovation: Living Proof, No Time to Die, Die Again Tomorrow, and Mother Knows Best. Peikoff holds a B.A. in Journalism from New York University and an M.S. in Bioethics from Columbia University. She lives in New Jersey with her husband and two young sons. Follow her on Twitter @KiraPeikoff.
Massive benefits of AI come with environmental and human costs. Can AI itself be part of the solution?
Generative AI has a large carbon footprint and other drawbacks. But AI can help mitigate its own harms—by plowing through mountains of data on extreme weather and human displacement.
The recent explosion of generative artificial intelligence tools like ChatGPT and Dall-E enabled anyone with internet access to harness AI’s power for enhanced productivity, creativity, and problem-solving. With their ever-improving capabilities and expanding user base, these tools proved useful across disciplines, from the creative to the scientific.
But beneath the technological wonders of human-like conversation and creative expression lies a dirty secret—an alarming environmental and human cost. AI has an immense carbon footprint. Systems like ChatGPT take months to train in high-powered data centers, which demand huge amounts of electricity, much of which is still generated with fossil fuels, as well as water for cooling. “One of the reasons why Open AI needs investments [to the tune of] $10 billion from Microsoft is because they need to pay for all of that computation,” says Kentaro Toyama, a computer scientist at the University of Michigan. There’s also an ecological toll from mining rare minerals required for hardware and infrastructure. This environmental exploitation pollutes land, triggers natural disasters and causes large-scale human displacement. Finally, for data labeling needed to train and correct AI algorithms, the Big Data industry employs cheap and exploitative labor, often from the Global South.
Generative AI tools are based on large language models (LLMs), with most well-known being various versions of GPT. LLMs can perform natural language processing, including translating, summarizing and answering questions. They use artificial neural networks, called deep learning or machine learning. Inspired by the human brain, neural networks are made of millions of artificial neurons. “The basic principles of neural networks were known even in the 1950s and 1960s,” Toyama says, “but it’s only now, with the tremendous amount of compute power that we have, as well as huge amounts of data, that it’s become possible to train generative AI models.”
Though there aren’t any official figures about the power consumption or emissions from data centers, experts estimate that they use one percent of global electricity—more than entire countries.
In recent months, much attention has gone to the transformative benefits of these technologies. But it’s important to consider that these remarkable advances may come at a price.
AI’s carbon footprint
In their latest annual report, 2023 Landscape: Confronting Tech Power, the AI Now Institute, an independent policy research entity focusing on the concentration of power in the tech industry, says: “The constant push for scale in artificial intelligence has led Big Tech firms to develop hugely energy-intensive computational models that optimize for ‘accuracy’—through increasingly large datasets and computationally intensive model training—over more efficient and sustainable alternatives.”
Though there aren’t any official figures about the power consumption or emissions from data centers, experts estimate that they use one percent of global electricity—more than entire countries. In 2019, Emma Strubell, then a graduate researcher at the University of Massachusetts Amherst, estimated that training a single LLM resulted in over 280,000 kg in CO2 emissions—an equivalent of driving almost 1.2 million km in a gas-powered car. A couple of years later, David Patterson, a computer scientist from the University of California Berkeley, and colleagues, estimated GPT-3’s carbon footprint at over 550,000 kg of CO2 In 2022, the tech company Hugging Face, estimated the carbon footprint of its own language model, BLOOM, as 25,000 kg in CO2 emissions. (BLOOM’s footprint is lower because Hugging Face uses renewable energy, but it doubled when other life-cycle processes like hardware manufacturing and use were added.)
Luckily, despite the growing size and numbers of data centers, their increasing energy demands and emissions have not kept pace proportionately—thanks to renewable energy sources and energy-efficient hardware.
But emissions don’t tell the full story.
AI’s hidden human cost
“If historical colonialism annexed territories, their resources, and the bodies that worked on them, data colonialism’s power grab is both simpler and deeper: the capture and control of human life itself through appropriating the data that can be extracted from it for profit.” So write Nick Couldry and Ulises Mejias, authors of the book The Costs of Connection.
The energy requirements, hardware manufacture and the cheap human labor behind AI systems disproportionately affect marginalized communities.
Technologies we use daily inexorably gather our data. “Human experience, potentially every layer and aspect of it, is becoming the target of profitable extraction,” Couldry and Meijas say. This feeds data capitalism, the economic model built on the extraction and commodification of data. While we are being dispossessed of our data, Big Tech commodifies it for their own benefit. This results in consolidation of power structures that reinforce existing race, gender, class and other inequalities.
“The political economy around tech and tech companies, and the development in advances in AI contribute to massive displacement and pollution, and significantly changes the built environment,” says technologist and activist Yeshi Milner, who founded Data For Black Lives (D4BL) to create measurable change in Black people’s lives using data. The energy requirements, hardware manufacture and the cheap human labor behind AI systems disproportionately affect marginalized communities.
AI’s recent explosive growth spiked the demand for manual, behind-the-scenes tasks, creating an industry described by Mary Gray and Siddharth Suri as “ghost work” in their book. This invisible human workforce that lies behind the “magic” of AI, is overworked and underpaid, and very often based in the Global South. For example, workers in Kenya who made less than $2 an hour, were the behind the mechanism that trained ChatGPT to properly talk about violence, hate speech and sexual abuse. And, according to an article in Analytics India Magazine, in some cases these workers may not have been paid at all, a case for wage theft. An exposé by the Washington Post describes “digital sweatshops” in the Philippines, where thousands of workers experience low wages, delays in payment, and wage theft by Remotasks, a platform owned by Scale AI, a $7 billion dollar American startup. Rights groups and labor researchers have flagged Scale AI as one company that flouts basic labor standards for workers abroad.
It is possible to draw a parallel with chattel slavery—the most significant economic event that continues to shape the modern world—to see the business structures that allow for the massive exploitation of people, Milner says. Back then, people got chocolate, sugar, cotton; today, they get generative AI tools. “What’s invisible through distance—because [tech companies] also control what we see—is the massive exploitation,” Milner says.
“At Data for Black Lives, we are less concerned with whether AI will become human…[W]e’re more concerned with the growing power of AI to decide who’s human and who’s not,” Milner says. As a decision-making force, AI becomes a “justifying factor for policies, practices, rules that not just reinforce, but are currently turning the clock back generations years on people’s civil and human rights.”
Ironically, AI plays an important role in mitigating its own harms—by plowing through mountains of data about weather changes, extreme weather events and human displacement.
Nuria Oliver, a computer scientist, and co-founder and vice-president of the European Laboratory of Learning and Intelligent Systems (ELLIS), says that instead of focusing on the hypothetical existential risks of today’s AI, we should talk about its real, tangible risks.
“Because AI is a transverse discipline that you can apply to any field [from education, journalism, medicine, to transportation and energy], it has a transformative power…and an exponential impact,” she says.
AI's accountability
“At the core of what we were arguing about data capitalism [is] a call to action to abolish Big Data,” says Milner. “Not to abolish data itself, but the power structures that concentrate [its] power in the hands of very few actors.”
A comprehensive AI Act currently negotiated in the European Parliament aims to rein Big Tech in. It plans to introduce a rating of AI tools based on the harms caused to humans, while being as technology-neutral as possible. That sets standards for safe, transparent, traceable, non-discriminatory, and environmentally friendly AI systems, overseen by people, not automation. The regulations also ask for transparency in the content used to train generative AIs, particularly with copyrighted data, and also disclosing that the content is AI-generated. “This European regulation is setting the example for other regions and countries in the world,” Oliver says. But, she adds, such transparencies are hard to achieve.
Google, for example, recently updated its privacy policy to say that anything on the public internet will be used as training data. “Obviously, technology companies have to respond to their economic interests, so their decisions are not necessarily going to be the best for society and for the environment,” Oliver says. “And that’s why we need strong research institutions and civil society institutions to push for actions.” ELLIS also advocates for data centers to be built in locations where the energy can be produced sustainably.
Ironically, AI plays an important role in mitigating its own harms—by plowing through mountains of data about weather changes, extreme weather events and human displacement. “The only way to make sense of this data is using machine learning methods,” Oliver says.
Milner believes that the best way to expose AI-caused systemic inequalities is through people's stories. “In these last five years, so much of our work [at D4BL] has been creating new datasets, new data tools, bringing the data to life. To show the harms but also to continue to reclaim it as a tool for social change and for political change.” This change, she adds, will depend on whose hands it is in.
DNA gathered from animal poop helps protect wildlife
Alida de Flamingh and her team are collecting elephant dung. It holds a trove of information about animal health, diet and genetic diversity.
On the savannah near the Botswana-Zimbabwe border, elephants grazed contentedly. Nearby, postdoctoral researcher Alida de Flamingh watched and waited. As the herd moved away, she went into action, collecting samples of elephant dung that she and other wildlife conservationists would study in the months to come. She pulled on gloves, took a swab, and ran it all over the still-warm, round blob of elephant poop.
Sequencing DNA from fecal matter is a safe, non-invasive way to track and ultimately help protect over 42,000 species currently threatened by extinction. Scientists are using this DNA to gain insights into wildlife health, genetic diversity and even the broader environment. Applied to elephants, chimpanzees, toucans and other species, it helps scientists determine the genetic diversity of groups and linkages with other groups. Such analysis can show changes in rates of inbreeding. Populations with greater genetic diversity adapt better to changes and environmental stressors than those with less diversity, thus reducing their risks of extinction, explains de Flamingh, a postdoctoral researcher at the University of Illinois Urbana-Champaign.
Analyzing fecal DNA also reveals information about an animal’s diet and health, and even nearby flora that is eaten. That information gives scientists broader insights into the ecosystem, and the findings are informing conservation initiatives. Examples include restoring or maintaining genetic connections among groups, ensuring access to certain foraging areas or increasing diversity in captive breeding programs.
Approximately 27 percent of mammals and 28 percent of all assessed species are close to dying out. The IUCN Red List of threatened species, simply called the Red List, is the world’s most comprehensive record of animals’ risk of extinction status. The more information scientists gather, the better their chances of reducing those risks. In Africa, populations of vertebrates declined 69 percent between 1970 and 2022, according to the World Wildlife Fund (WWF).
“We put on sterile gloves and use a sterile swab to collect wet mucus and materials from the outside of the dung ball,” says Alida de Flamingh, a postdoctoral researcher at the University of Illinois Urbana-Champaign.
“When people talk about species, they often talk about ecosystems, but they often overlook genetic diversity,” says Christina Hvilsom, senior geneticist at the Copenhagen Zoo. “It’s easy to count (individuals) to assess whether the population size is increasing or decreasing, but diversity isn’t something we can see with our bare eyes. Yet, it’s actually the foundation for the species and populations.” DNA analysis can provide this critical information.
Assessing elephants’ health
“Africa’s elephant populations are facing unprecedented threats,” says de Flamingh, the postdoc, who has studied them since 2009. Challenges include ivory poaching, habitat destruction and smaller, more fragmented habitats that result in smaller mating pools with less genetic diversity. Additionally, de Flamingh studies the microbial communities living on and in elephants – their microbiomes – looking for parasites or dangerous microbes.
Approximately 415,000 elephants inhabit Africa today, but de Flamingh says the number would be four times higher without these challenges. The IUCN Red List reports African savannah elephants are endangered and African forest elephants are critically endangered. Elephants support ecosystem biodiversity by clearing paths that help other species travel. Their very footprints create small puddles that can host smaller organisms such as tadpoles. Elephants are often described as ecosystems’ engineers, so if they disappear, the rest of the ecosystem will suffer too.
There’s a process to collecting elephant feces. “We put on sterile gloves (which we change for each sample) and use a sterile swab to collect wet mucus and materials from the outside of the dung ball,” says de Flamingh. They rub a sample about the size of a U.S. quarter onto a paper card embedded with DNA preservation technology. Each card is air dried and stored in a packet of desiccant to prevent mold growth. This way, samples can be stored at room temperature indefinitely without the DNA degrading.
Earlier methods required collecting dung in bags, which needed either refrigeration or the addition of preservatives, or the riskier alternative of tranquilizing the animals before approaching them to draw blood samples. The ability to collect and sequence the DNA made things much easier and safer.
“Our research provides a way to assess elephant health without having to physically interact with elephants,” de Flamingh emphasizes. “We also keep track of the GPS coordinates of each sample so that we can create a map of the sampling locations,” she adds. That helps researchers correlate elephants’ health with geographic areas and their conditions.
Although de Flamingh works with elephants in the wild, the contributions of zoos in the United States and collaborations in South Africa (notably the late Professor Rudi van Aarde and the Conservation Ecology Research Unit at the University of Pretoria) were key in studying this method to ensure it worked, she points out.
Protecting chimpanzees
Genetic work with chimpanzees began about a decade ago. Hvilsom and her group at the Copenhagen Zoo analyzed DNA from nearly 1,000 fecal samples collected between 2003 and 2018 by a team of international researchers. The goal was to assess the status of the West African subspecies, which is critically endangered after rapid population declines. Of the four subspecies of chimpanzees, the West African subspecies is considered the most at-risk.
In total, the WWF estimates the numbers of chimpanzees inhabiting Africa’s forests and savannah woodlands at between 173,000 and 300,000. Poaching, disease and human-caused changes to their lands are their major risks.
By analyzing genetics obtained from fecal samples, Hvilsom estimated the chimpanzees’ population, ascertained their family relationships and mapped their migration routes.
“One of the threats is mining near the Nimba Mountains in Guinea,” a stronghold for the West African subspecies, Hvilsom says. The Nimba Mountains are a UNESCO World Heritage Site, but they are rich in iron ore, which is used to make the steel that is vital to the Asian construction boom. As she and colleagues wrote in a recent paper, “Many extractive industries are currently developing projects in chimpanzee habitat.”
Analyzing DNA allows researchers to identify individual chimpanzees more accurately than simply observing them, she says. Normally, field researchers would install cameras and manually inspect each picture to determine how many chimpanzees were in an area. But, Hvilsom says, “That’s very tricky. Chimpanzees move a lot and are fast, so it’s difficult to get clear pictures. Often, they find and destroy the cameras. Also, they live in large areas, so you need a lot of cameras.”
By analyzing genetics obtained from fecal samples, Hvilsom estimated the chimpanzees’ population, ascertained their family relationships and mapped their migration routes based upon DNA comparisons with other chimpanzee groups. The mining companies and builders are using this information to locate future roads where they won’t disrupt migration – a more effective solution than trying to build artificial corridors for wildlife.
“The current route cuts off communities of chimpanzees,” Hvilsom elaborates. That effectively prevents young adult chimps from joining other groups when the time comes, eventually reducing the currently-high levels of genetic diversity.
“The mining company helped pay for the genetics work,” Hvilsom says, “as part of its obligation to assess and monitor biodiversity and the effect of the mining in the area.”
Of 50 toucan subspecies, 11 are threatened or near-threatened with extinction because of deforestation and poaching.
Identifying toucan families
Feces aren't the only substance researchers draw DNA samples from. Jeffrey Coleman, a Ph.D. candidate at the University of Texas at Austin relies on blood tests for studying the genetic diversity of toucans---birds species native to Central America and nearby regions. They live in the jungles, where they hop among branches, snip fruit from trees, toss it in the air and catch it with their large beaks. “Toucans are beautiful, charismatic birds that are really important to the ecosystem,” says Coleman.
Of their 50 subspecies, 11 are threatened or near-threatened with extinction because of deforestation and poaching. “When people see these aesthetically pleasing birds, they’re motivated to care about conservation practices,” he points out.
Coleman works with the Dallas World Aquarium and its partner zoos to analyze DNA from blood draws, using it to identify which toucans are related and how closely. His goal is to use science to improve the genetic diversity among toucan offspring.
Specifically, he’s looking at sections of the genome of captive birds in which the nucleotides repeat multiple times, such as AGATAGATAGAT. Called microsatellites, these consecutively-repeating sections can be passed from parents to children, helping scientists identify parent-child and sibling-sibling relationships. “That allows you to make strategic decisions about how to pair (captive) individuals for mating...to avoid inbreeding,” Coleman says.
Jeffrey Coleman is studying the microsatellites inside the toucan genomes.
Courtesy Jeffrey Coleman
The alternative is to use a type of analysis that looks for a single DNA building block – a nucleotide – that differs in a given sequence. Called single nucleotide polymorphisms (SNPs, pronounced “snips”), they are very common and very accurate. Coleman says they are better than microsatellites for some uses. But scientists have already developed a large body of microsatellite data from multiple species, so microsatellites can shed more insights on relations.
Regardless of whether conservation programs use SNPs or microsatellites to guide captive breeding efforts, the goal is to help them build genetically diverse populations that eventually may supplement endangered populations in the wild. “The hope is that the ecosystem will be stable enough and that the populations (once reintroduced into the wild) will be able to survive and thrive,” says Coleman. History knows some good examples of captive breeding success.
The California condor, which had a total population of 27 in 1987, when the last wild birds were captured, is one of them. A captive breeding program boosted their numbers to 561 by the end of 2022. Of those, 347 of those are in the wild, according to the National Park Service.
Conservationists hope that their work on animals’ genetic diversity will help preserve and restore endangered species in captivity and the wild. DNA analysis is crucial to both types of efforts. The ability to apply genome sequencing to wildlife conservation brings a new level of accuracy that helps protect species and gives fresh insights that observation alone can’t provide.
“A lot of species are threatened,” Coleman says. “I hope this research will be a resource people can use to get more information on longer-term genealogies and different populations.”

