Food Poisoning Outbreaks Are Still A Problem. Powerful Tech Is Fighting Back.

The latest tools, like whole genome sequencing, are allowing food outbreaks to be identified and stopped more quickly.
With the pandemic at the forefront of everyone's minds, many people have wondered if food could be a source of coronavirus transmission. Luckily, that "seems unlikely," according to the CDC, but foodborne illnesses do still sicken a whopping 48 million people per year.
Whole genome sequencing is like "going from an eight-bit image—maybe like what you would see in Minecraft—to a high definition image."
In normal times, when there isn't a historic global health crisis infecting millions and affecting the lives of billions, foodborne outbreaks are real and frightening, potentially deadly, and can cause widespread fear of particular foods. Think of Romaine lettuce spreading E. coli last year— an outbreak that infected more than 500 people and killed eight—or peanut butter spreading salmonella in 2008, which infected 167 people.
The technologies available to detect and prevent the next foodborne disease outbreak have improved greatly over the past 30-plus years, particularly during the past decade, and better, more nimble technologies are being developed, according to experts in government, academia, and private industry. The key to advancing detection of harmful foodborne pathogens, they say, is increasing speed and portability of detection, and the precision of that detection.
Getting to Rapid Results
Researchers at Purdue University have recently developed a lateral flow assay that, with the help of a laser, can detect toxins and pathogenic E. coli. Lateral flow assays are cheap and easy to use; a good example is a home pregnancy test. You place a liquid or liquefied sample on a piece of paper designed to detect a single substance and soon after you get the results in the form of a colored line: yes or no.
"They're a great portable tool for us for food contaminant detection," says Carmen Gondhalekar, a fifth-year biomedical engineering graduate student at Purdue. "But one of the areas where paper-based lateral flow assays could use improvement is in multiplexing capability and their sensitivity."
J. Paul Robinson, a professor in Purdue's Colleges of Veterinary Medicine and Engineering, and Gondhalekar's advisor, agrees. "One of the fundamental problems that we have in detection is that it is hard to identify pathogens in complex samples," he says.
When it comes to foodborne disease outbreaks, you don't always know what substance you're looking for, so an assay made to detect only a single substance isn't always effective. The goal of the project at Purdue is to make assays that can detect multiple substances at once.
These assays would be more complex than a pregnancy test. As detailed in Gondhalekar's recent paper, a laser pulse helps create a spectral signal from the sample on the assay paper, and the spectral signal is then used to determine if any unique wavelengths associated with one of several toxins or pathogens are present in the sample. Though the handheld technology has yet to be built, the idea is that the results would be given on the spot. So someone in the field trying to track the source of a Salmonella infection could, for instance, put a suspected lettuce sample on the assay and see if it has the pathogen on it.
"What our technology is designed to do is to give you a rapid assessment of the sample," says Robinson. "The goal here is speed."
Seeing the Pathogen in "High-Def"
"One in six Americans will get a foodborne illness every year," according to Dr. Heather Carleton, a microbiologist at the Centers for Disease Control and Prevention's Enteric Diseases Laboratory Branch. But not every foodborne outbreak makes the news. In 2017 alone, the CDC monitored between 18 and 37 foodborne poison clusters per week and investigated 200 multi-state clusters. Hardboiled eggs, ground beef, chopped salad kits, raw oysters, frozen tuna, and pre-cut melon are just a taste of the foods that were investigated last year for different strains of listeria, salmonella, and E. coli.
At the heart of the CDC investigations is PulseNet, a national network of laboratories that uses DNA fingerprinting to detect outbreaks at local and regional levels. This is how it works: When a patient gets sick—with symptoms like vomiting and fever, for instance—they will go to a hospital or clinic for treatment. Since we're talking about foodborne illnesses, a clinician will likely take a stool sample from the patient and send it off to a laboratory to see if there is a foodborne pathogen, like salmonella, E. Coli, or another one. If it does contain a potentially harmful pathogen, then a bacterial isolate of that identified sample is sent to a regional public health lab so that whole genome sequencing can be performed.
Whole genome sequencing can differentiate "virtually all" strains of foodborne pathogens, no matter the species, according to the FDA.
Whole genome sequencing is a method for reading the entire genome of a bacterial isolate (or from any organism, for that matter). Instead of working with a couple dozen data points, now you're working with millions of base pairs. Carleton likes to describe it as "going from an eight-bit image—maybe like what you would see in Minecraft—to a high definition image," she says. "It's really an evolution of how we detect foodborne illnesses and identify outbreaks."
If the bacterial isolate matches another in the CDC's database, this means there could be a potential outbreak and an investigation may be started, with the goal of tracking the pathogen to its source.
Whole genome sequencing has been a relatively recent shift in foodborne disease detection. For more than 20 years, the standard technique for analyzing pathogens in foodborne disease outbreaks was pulsed-field gel electrophoresis. This method creates a DNA fingerprint for each sample in the form of a pattern of about 15-30 "bands," with each band representing a piece of DNA. Researchers like Carleton can use this fingerprint to see if two samples are from the same bacteria. The problem is that 15-30 bands are not enough to differentiate all isolates. Some isolates whose bands look very similar may actually come from different sources and some whose bands look different may be from the same source. But if you can see the entire DNA fingerprint, then you don't have that issue. That's where whole genome sequencing comes in.
Although the PulseNet team had piloted whole genome sequencing as early as 2013, it wasn't until July of last year that the transition to using whole genome sequencing for all pathogens was complete. Though whole genome sequencing requires far more computing power to generate, analyze, and compare those millions of data points, the payoff is huge.
Stopping Outbreaks Sooner
The U.S. Food and Drug Administration (FDA) acquired their first whole genome sequencers in 2008, according to Dr. Eric Brown, the Director of the Division of Microbiology in the FDA's Office of Regulatory Science. Since then, through their GenomeTrakr program, a network of more than 60 domestic and international labs, the FDA has sequenced and publicly shared more than 400,000 isolates. "The impact of what whole genome sequencing could do to resolve a foodborne outbreak event was no less impactful than when NASA turned on the Hubble Telescope for the first time," says Brown.
Whole genome sequencing has helped identify strains of Salmonella that prior methods were unable to differentiate. In fact, whole genome sequencing can differentiate "virtually all" strains of foodborne pathogens, no matter the species, according to the FDA. This means it takes fewer clinical cases—fewer sick people—to detect and end an outbreak.
And perhaps the largest benefit of whole genome sequencing is that these detailed sequences—the millions of base pairs—can imply geographic location. The genomic information of bacterial strains can be different depending on the area of the country, helping these public health agencies eventually track the source of outbreaks—a restaurant, a farm, a food-processing center.
Coming Soon: "Lab in a Backpack"
Now that whole genome sequencing has become the go-to technology of choice for analyzing foodborne pathogens, the next step is making the process nimbler and more portable. Putting "the lab in a backpack," as Brown says.
The CDC's Carleton agrees. "Right now, the sequencer we use is a fairly big box that weighs about 60 pounds," she says. "We can't take it into the field."
A company called Oxford Nanopore Technologies is developing handheld sequencers. Their devices are meant to "enable the sequencing of anything by anyone anywhere," according to Dan Turner, the VP of Applications at Oxford Nanopore.
"The sooner that we can see linkages…the sooner the FDA gets in action to mitigate the problem and put in some kind of preventative control."
"Right now, sequencing is very much something that is done by people in white coats in laboratories that are set up for that purpose," says Turner. Oxford Nanopore would like to create a new, democratized paradigm.
The FDA is currently testing these types of portable sequencers. "We're very excited about it. We've done some pilots, to be able to do that sequencing in the field. To actually do it at a pond, at a river, at a canal. To do it on site right there," says Brown. "This, of course, is huge because it means we can have real-time sequencing capability to stay in step with an actual laboratory investigation in the field."
"The timeliness of this information is critical," says Marc Allard, a senior biomedical research officer and Brown's colleague at the FDA. "The sooner that we can see linkages…the sooner the FDA gets in action to mitigate the problem and put in some kind of preventative control."
At the moment, the world is rightly focused on COVID-19. But as the danger of one virus subsides, it's only a matter of time before another pathogen strikes. Hopefully, with new and advancing technology like whole genome sequencing, we can stop the next deadly outbreak before it really gets going.
A robot server, controlled remotely by a disabled worker, delivers drinks to patrons at the DAWN cafe in Tokyo.
A sleek, four-foot tall white robot glides across a cafe storefront in Tokyo’s Nihonbashi district, holding a two-tiered serving tray full of tea sandwiches and pastries. The cafe’s patrons smile and say thanks as they take the tray—but it’s not the robot they’re thanking. Instead, the patrons are talking to the person controlling the robot—a restaurant employee who operates the avatar from the comfort of their home.
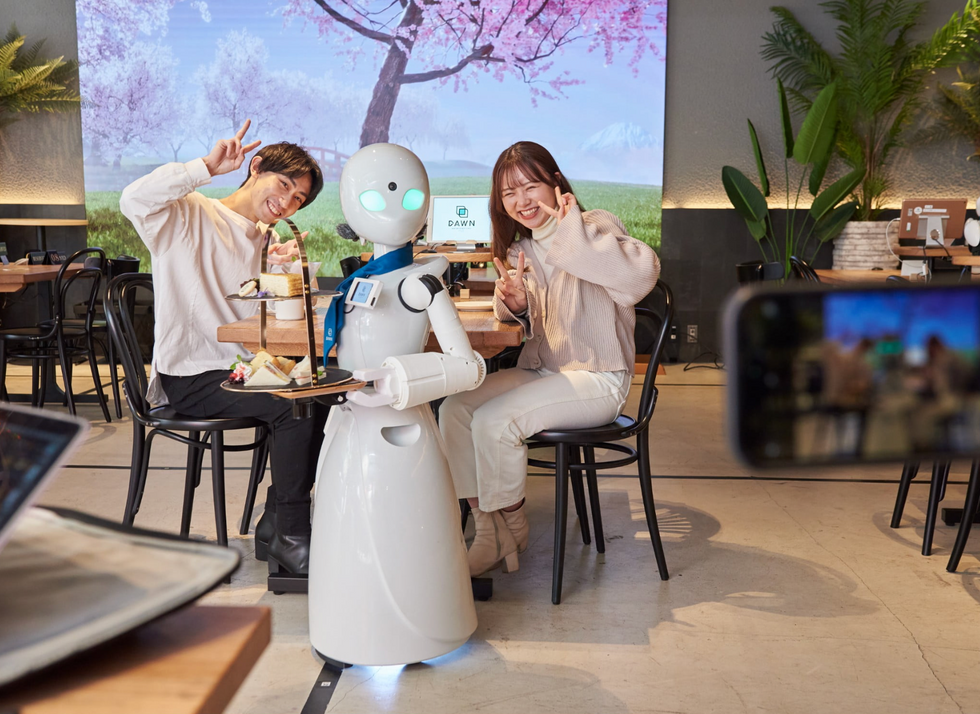
It’s a typical scene at DAWN, short for Diverse Avatar Working Network—a cafe that launched in Tokyo six years ago as an experimental pop-up and quickly became an overnight success. Today, the cafe is a permanent fixture in Nihonbashi, staffing roughly 60 remote workers who control the robots remotely and communicate to customers via a built-in microphone.
More than just a creative idea, however, DAWN is being hailed as a life-changing opportunity. The workers who control the robots remotely (known as “pilots”) all have disabilities that limit their ability to move around freely and travel outside their homes. Worldwide, an estimated 16 percent of the global population lives with a significant disability—and according to the World Health Organization, these disabilities give rise to other problems, such as exclusion from education, unemployment, and poverty.
These are all problems that Kentaro Yoshifuji, founder and CEO of Ory Laboratory, which supplies the robot servers at DAWN, is looking to correct. Yoshifuji, who was bedridden for several years in high school due to an undisclosed health problem, launched the company to help enable people who are house-bound or bedridden to more fully participate in society, as well as end the loneliness, isolation, and feelings of worthlessness that can sometimes go hand-in-hand with being disabled.
“It’s heartbreaking to think that [people with disabilities] feel they are a burden to society, or that they fear their families suffer by caring for them,” said Yoshifuji in an interview in 2020. “We are dedicating ourselves to providing workable, technology-based solutions. That is our purpose.”
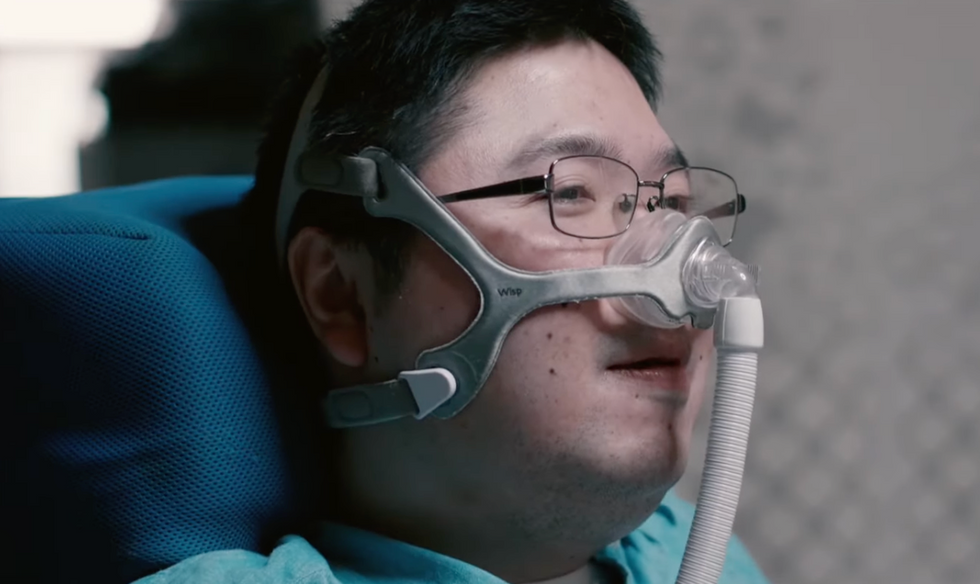
Shota, Kuwahara, a DAWN employee with muscular dystrophy, agrees. "There are many difficulties in my daily life, but I believe my life has a purpose and is not being wasted," he says. "Being useful, able to help other people, even feeling needed by others, is so motivational."
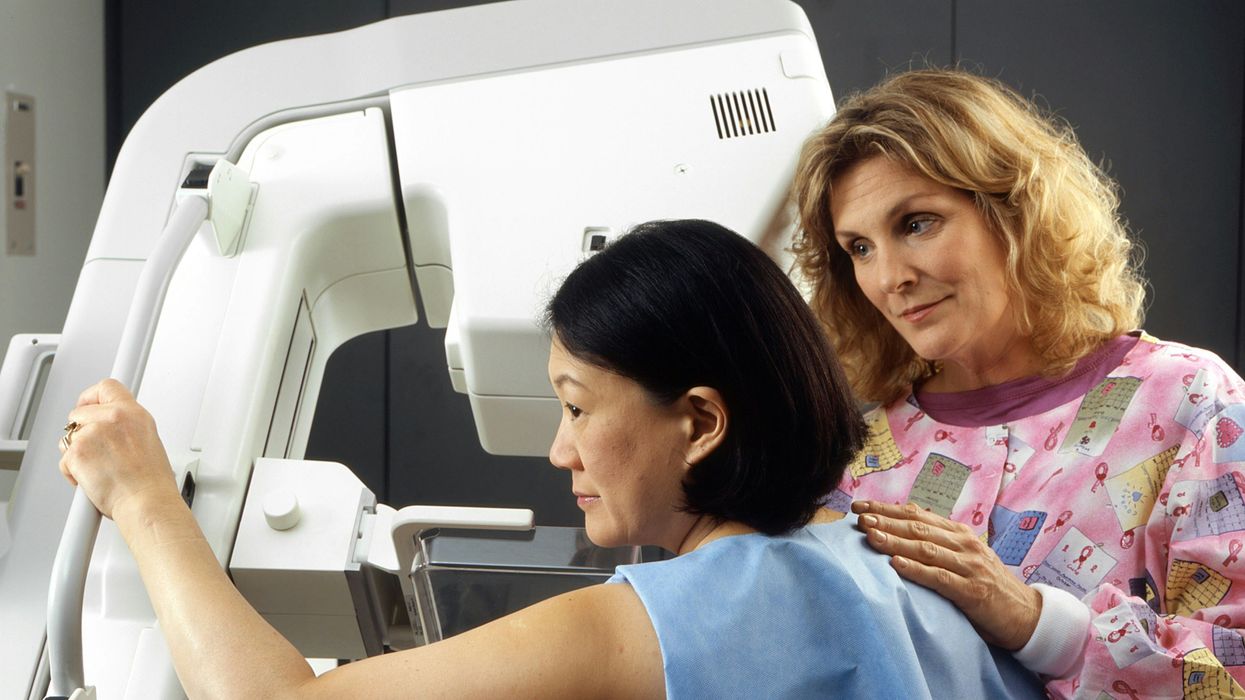
A woman receives a mammogram, which can detect the presence of tumors in a patient's breast.
When a patient is diagnosed with early-stage breast cancer, having surgery to remove the tumor is considered the standard of care. But what happens when a patient can’t have surgery?
Whether it’s due to high blood pressure, advanced age, heart issues, or other reasons, some breast cancer patients don’t qualify for a lumpectomy—one of the most common treatment options for early-stage breast cancer. A lumpectomy surgically removes the tumor while keeping the patient’s breast intact, while a mastectomy removes the entire breast and nearby lymph nodes.
Fortunately, a new technique called cryoablation is now available for breast cancer patients who either aren’t candidates for surgery or don’t feel comfortable undergoing a surgical procedure. With cryoablation, doctors use an ultrasound or CT scan to locate any tumors inside the patient’s breast. They then insert small, needle-like probes into the patient's breast which create an “ice ball” that surrounds the tumor and kills the cancer cells.
Cryoablation has been used for decades to treat cancers of the kidneys and liver—but only in the past few years have doctors been able to use the procedure to treat breast cancer patients. And while clinical trials have shown that cryoablation works for tumors smaller than 1.5 centimeters, a recent clinical trial at Memorial Sloan Kettering Cancer Center in New York has shown that it can work for larger tumors, too.
In this study, doctors performed cryoablation on patients whose tumors were, on average, 2.5 centimeters. The cryoablation procedure lasted for about 30 minutes, and patients were able to go home on the same day following treatment. Doctors then followed up with the patients after 16 months. In the follow-up, doctors found the recurrence rate for tumors after using cryoablation was only 10 percent.
For patients who don’t qualify for surgery, radiation and hormonal therapy is typically used to treat tumors. However, said Yolanda Brice, M.D., an interventional radiologist at Memorial Sloan Kettering Cancer Center, “when treated with only radiation and hormonal therapy, the tumors will eventually return.” Cryotherapy, Brice said, could be a more effective way to treat cancer for patients who can’t have surgery.
“The fact that we only saw a 10 percent recurrence rate in our study is incredibly promising,” she said.