With this new technology, hospitals and pharmacies could make vaccines and medicines onsite
Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.

New research focuses on methods that could change medicine-making worldwide. The scientists propose bursting cells open, removing their DNA and using the cellular gears inside to make therapies.
Most modern biopharmaceutical medicines are produced by workhorse cells—typically bacterial but sometimes mammalian. The cells receive the synthesizing instructions on a snippet of a genetic code, which they incorporate into their DNA. The cellular machinery—ribosomes, RNAs, polymerases, and other compounds—read and use these instructions to build the medicinal molecules, which are harvested and administered to patients.
Although a staple of modern pharma, this process is complex and expensive. One must first insert the DNA instructions into the cells, which they may or may not uptake. One then must grow the cells, keeping them alive and well, so that they produce the required therapeutics, which then must be isolated and purified. To make this at scale requires massive bioreactors and big factories from where the drugs are distributed—and may take a while to arrive where they’re needed. “The pandemic showed us that this method is slow and cumbersome,” says Govind Rao, professor of biochemical engineering who directs the Center for Advanced Sensor Technology at the University of Maryland, Baltimore County (UMBC). “We need better methods that can work faster and can work locally where an outbreak is happening.”
Rao and his team of collaborators, which spans multiple research institutions, believe they have a better approach that may change medicine-making worldwide. They suggest forgoing the concept of using living cells as medicine-producers. Instead, they propose breaking the cells and using the remaining cellular gears for assembling the therapeutic compounds. Instead of inserting the DNA into living cells, the team burst them open, and removed their DNA altogether. Yet, the residual molecular machinery of ribosomes, polymerases and other cogwheels still functioned the way it would in a cell. “Now if you drop your DNA drug-making instructions into that soup, this machinery starts making what you need,” Rao explains. “And because you're no longer worrying about living cells, it becomes much simpler and more efficient.” The collaborators detail their cell-free protein synthesis or CFPS method in their recent paper published in preprint BioAxiv.
While CFPS does not use living cells, it still needs the basic building blocks to assemble proteins from—such as amino acids, nucleotides and certain types of enzymes. These are regularly added into this “soup” to keep the molecular factory chugging. “We just mix everything in as a batch and we let it integrate,” says James Robert Swartz, professor of chemical engineering and bioengineering at Stanford University and co-author of the paper. “And we make sure that we provide enough oxygen.” Rao likens the process to making milk from milk powder.
For a variety of reasons—from the field’s general inertia to regulatory approval hurdles—the method hasn’t become mainstream. The pandemic rekindled interest in medicines that can be made quickly and easily, so it drew more attention to the technology.
The idea of a cell-free protein synthesis is older than one might think. Swartz first experimented with it around 1997, when he was a chemical engineer at Genentech. While working on engineering bacteria to make pharmaceuticals, he discovered that there was a limit to what E. coli cells, the workhorse darling of pharma, could do. For example, it couldn’t grow and properly fold some complex proteins. “We tried many genetic engineering approaches, many fermentation, development, and environmental control approaches,” Swartz recalls—to no avail.
“The organism had its own agenda,” he quips. “And because everything was happening within the organism, we just couldn't really change those conditions very easily. Some of them we couldn’t change at all—we didn’t have control.”
It was out of frustration with the defiant bacteria that a new idea took hold. Could the cells be opened instead, so that the protein-forming reactions could be influenced more easily? “Obviously, we’d lose the ability for them to reproduce,” Swartz says. But that also meant that they no longer needed to keep the cells alive and could focus on making the specific reactions happen. “We could take the catalysts, the enzymes, and the more complex catalysts and activate them, make them work together, much as they would in a living cell, but the way we wanted.”
In 1998, Swartz joined Stanford, and began perfecting the biochemistry of the cell-free method, identifying the reactions he wanted to foster and stopping those he didn’t want. He managed to make the idea work, but for a variety of reasons—from the field’s general inertia to regulatory approval hurdles—the method hasn’t become mainstream. The pandemic rekindled interest in medicines that can be made quickly and easily, so it drew more attention to the technology. For their BioArxiv paper, the team tested the method by growing a specific antiviral protein called griffithsin.
First identified by Barry O’Keefe at National Cancer Institute over a decade ago, griffithsin is an antiviral known to interfere with many viruses’ ability to enter cells—including HIV, SARS, SARS-CoV-2, MERS and others. Originally isolated from the red algae Griffithsia, it works differently from antibodies and antibody cocktails.
Most antiviral medicines tend to target the specific receptors that viruses use to gain entry to the cells they infect. For example, SARS-CoV-2 uses the infamous spike protein to latch onto the ACE2 receptor of mammalian cells. The antibodies or other antiviral molecules stick to the spike protein, shutting off its ability to cling onto the ACE2 receptors. Unfortunately, the spike proteins mutate very often, so the medicines lose their potency. On the contrary, griffithsin has the ability to cling to the different parts of viral shells called capsids—namely to the molecules of mannose, a type of sugar. That extra stuff, glued all around the capsid like dead weight, makes it impossible for the virus to squeeze into the cell.
“Every time we have a vaccine or an antibody against a specific SARS-CoV-2 strain, that strain then mutates and so you lose efficacy,” Rao explains. “But griffithsin molecules glom onto the viral capsid, so the capsid essentially becomes a sticky mess and can’t enter the cell.” Mannose molecules also don’t mutate as easily as viruses’ receptors, so griffithsin-based antivirals do not have to be constantly updated. And because mannose molecules are found on many viruses’ capsids, it makes griffithsin “a universal neutralizer,” Rao explains.
“When griffithsin was discovered, we recognized that it held a lot of promise as a potential antiviral agent,” O’Keefe says. In 2010, he published a paper about griffithsin efficacy in neutralizing viruses of the corona family—after the first SARS outbreak in the early 2000s, the scientific community was interested in such antivirals. Yet, griffithsin is still not available as an off-the-shelf product. So during the Covid pandemic, the team experimented with synthesizing griffithsin using the cell-free production method. They were able to generate potent griffithsin in less than 24 hours without having to grow living cells.
The antiviral protein isn't the only type of medicine that can be made cell-free. The proteins needed for vaccine production could also be made the same way. “Such portable, on-demand drug manufacturing platforms can produce antiviral proteins within hours, making them ideal for combating future pandemics,” Rao says. “We would be able to stop the pandemic before it spreads.”
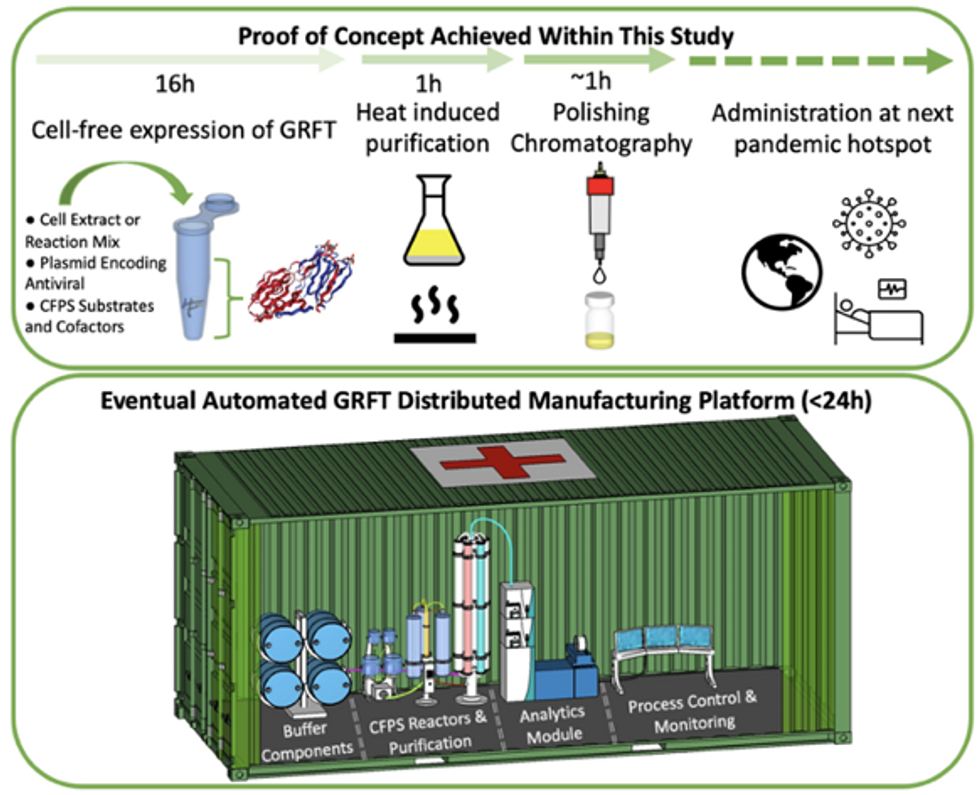
Top: Describes the process used in the study. Bottom: Describes how the new medicines and vaccines could be made at the site of a future viral outbreak.
Image courtesy of Rao and team, sourced from An approach to rapid distributed manufacturing of broad spectrumanti-viral griffithsin using cell-free systems to mitigate pandemics.
Rao’s idea is to perfect the technology to the point that any hospital or pharmacy can load up the media containing molecular factories, mix up the required amino acids, nucleotides and enzymes, and harvest the meds within hours. That will allow making medicines onsite and on demand. “That would be a self-contained production unit, so that you could just ship the production wherever the pandemic is breaking out,” says Swartz.
These units and the meds they produce, will, of course, have to undergo rigorous testing. “The biggest hurdles will be validating these against conventional technology,” Rao says. The biotech industry is risk-averse and prefers the familiar methods. But if this approach works, it may go beyond emergency situations and revolutionize the medicine-making paradigm even outside hospitals and pharmacies. Rao hopes that someday the method might become so mainstream that people may be able to buy and operate such reactors at home. “You can imagine a diabetic patient making insulin that way, or some other drugs,” Rao says. It would work not unlike making baby formula from the mere white powder. Just add water—and some oxygen, too.
Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.
Jamie Rettinger with his now fiance Amie Purnel-Davis, who helped him through the clinical trial.
Jamie Rettinger was still in his thirties when he first noticed a tiny streak of brown running through the thumbnail of his right hand. It slowly grew wider and the skin underneath began to deteriorate before he went to a local dermatologist in 2013. The doctor thought it was a wart and tried scooping it out, treating the affected area for three years before finally removing the nail bed and sending it off to a pathology lab for analysis.
"I have some bad news for you; what we removed was a five-millimeter melanoma, a cancerous tumor that often spreads," Jamie recalls being told on his return visit. "I'd never heard of cancer coming through a thumbnail," he says. None of his doctors had ever mentioned it either. "I just thought I was being treated for a wart." But nothing was healing and it continued to bleed.
A few months later a surgeon amputated the top half of his thumb. Lymph node biopsy tested negative for spread of the cancer and when the bandages finally came off, Jamie thought his medical issues were resolved.
Melanoma is the deadliest form of skin cancer. About 85,000 people are diagnosed with it each year in the U.S. and more than 8,000 die of the cancer when it spreads to other parts of the body, according to the Centers for Disease Control and Prevention (CDC).
There are two peaks in diagnosis of melanoma; one is in younger women ages 30-40 and often is tied to past use of tanning beds; the second is older men 60+ and is related to outdoor activity from farming to sports. Light-skinned people have a twenty-times greater risk of melanoma than do people with dark skin.
"When I graduated from medical school, in 2005, melanoma was a death sentence" --Diwakar Davar.
Jamie had a follow up PET scan about six months after his surgery. A suspicious spot on his lung led to a biopsy that came back positive for melanoma. The cancer had spread. Treatment with a monoclonal antibody (nivolumab/Opdivo®) didn't prove effective and he was referred to the UPMC Hillman Cancer Center in Pittsburgh, a four-hour drive from his home in western Ohio.
An alternative monoclonal antibody treatment brought on such bad side effects, diarrhea as often as 15 times a day, that it took more than a week of hospitalization to stabilize his condition. The only options left were experimental approaches in clinical trials.
Early research
"When I graduated from medical school, in 2005, melanoma was a death sentence" with a cure rate in the single digits, says Diwakar Davar, 39, an oncologist at UPMC Hillman Cancer Center who specializes in skin cancer. That began to change in 2010 with introduction of the first immunotherapies, monoclonal antibodies, to treat cancer. The antibodies attach to PD-1, a receptor on the surface of T cells of the immune system and on cancer cells. Antibody treatment boosted the melanoma cure rate to about 30 percent. The search was on to understand why some people responded to these drugs and others did not.
At the same time, there was a growing understanding of the role that bacteria in the gut, the gut microbiome, plays in helping to train and maintain the function of the body's various immune cells. Perhaps the bacteria also plays a role in shaping the immune response to cancer therapy.
One clue came from genetically identical mice. Animals ordered from different suppliers sometimes responded differently to the experiments being performed. That difference was traced to different compositions of their gut microbiome; transferring the microbiome from one animal to another in a process known as fecal transplant (FMT) could change their responses to disease or treatment.
When researchers looked at humans, they found that the patients who responded well to immunotherapies had a gut microbiome that looked like healthy normal folks, but patients who didn't respond had missing or reduced strains of bacteria.
Davar and his team knew that FMT had a very successful cure rate in treating the gut dysbiosis of Clostridioides difficile, a persistant intestinal infection, and they wondered if a fecal transplant from a patient who had responded well to cancer immunotherapy treatment might improve the cure rate of patients who did not originally respond to immunotherapies for melanoma.
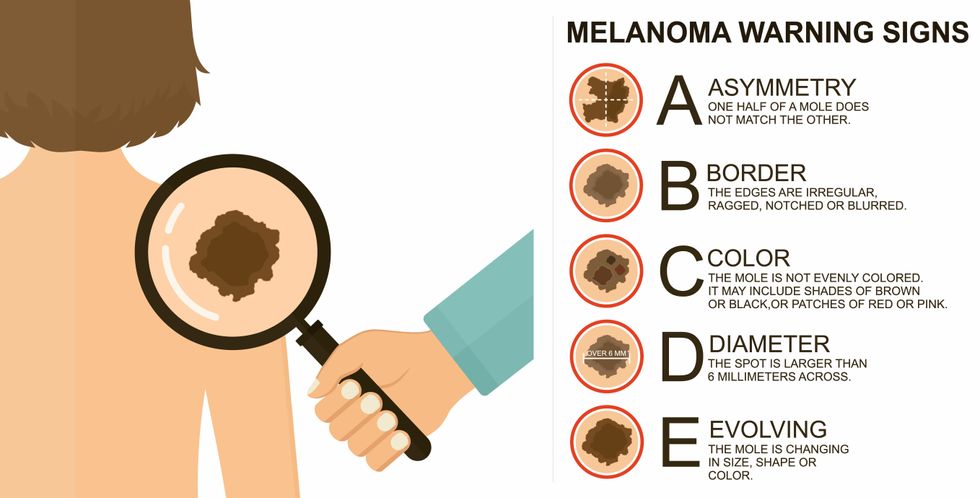
The ABCDE of melanoma detection
Adobe Stock
Clinical trial
"It was pretty weird, I was totally blasted away. Who had thought of this?" Jamie first thought when the hypothesis was explained to him. But Davar's explanation that the procedure might restore some of the beneficial bacterial his gut was lacking, convinced him to try. He quickly signed on in October 2018 to be the first person in the clinical trial.
Fecal donations go through the same safety procedures of screening for and inactivating diseases that are used in processing blood donations to make them safe for transfusion. The procedure itself uses a standard hollow colonoscope designed to screen for colon cancer and remove polyps. The transplant is inserted through the center of the flexible tube.
Most patients are sedated for procedures that use a colonoscope but Jamie doesn't respond to those drugs: "You can't knock me out. I was watching them on the TV going up my own butt. It was kind of unreal at that point," he says. "There were about twelve people in there watching because no one had seen this done before."
A test two weeks after the procedure showed that the FMT had engrafted and the once-missing bacteria were thriving in his gut. More importantly, his body was responding to another monoclonal antibody (pembrolizumab/Keytruda®) and signs of melanoma began to shrink. Every three months he made the four-hour drive from home to Pittsburgh for six rounds of treatment with the antibody drug.
"We were very, very lucky that the first patient had a great response," says Davar. "It allowed us to believe that even though we failed with the next six, we were on the right track. We just needed to tweak the [fecal] cocktail a little better" and enroll patients in the study who had less aggressive tumor growth and were likely to live long enough to complete the extensive rounds of therapy. Six of 15 patients responded positively in the pilot clinical trial that was published in the journal Science.
Davar believes they are beginning to understand the biological mechanisms of why some patients initially do not respond to immunotherapy but later can with a FMT. It is tied to the background level of inflammation produced by the interaction between the microbiome and the immune system. That paper is not yet published.
Surviving cancer
It has been almost a year since the last in his series of cancer treatments and Jamie has no measurable disease. He is cautiously optimistic that his cancer is not simply in remission but is gone for good. "I'm still scared every time I get my scans, because you don't know whether it is going to come back or not. And to realize that it is something that is totally out of my control."
"It was hard for me to regain trust" after being misdiagnosed and mistreated by several doctors he says. But his experience at Hillman helped to restore that trust "because they were interested in me, not just fixing the problem."
He is grateful for the support provided by family and friends over the last eight years. After a pause and a sigh, the ruggedly built 47-year-old says, "If everyone else was dead in my family, I probably wouldn't have been able to do it."
"I never hesitated to ask a question and I never hesitated to get a second opinion." But Jamie acknowledges the experience has made him more aware of the need for regular preventive medical care and a primary care physician. That person might have caught his melanoma at an earlier stage when it was easier to treat.
Davar continues to work on clinical studies to optimize this treatment approach. Perhaps down the road, screening the microbiome will be standard for melanoma and other cancers prior to using immunotherapies, and the FMT will be as simple as swallowing a handful of freeze-dried capsules off the shelf rather than through a colonoscopy. Earlier this year, the Food and Drug Administration approved the first oral fecal microbiota product for C. difficile, hopefully paving the way for more.
An older version of this hit article was first published on May 18, 2021
All organisms can repair damaged tissue, but none do it better than salamanders and newts. A surprising area of science could tell us how they manage this feat - and perhaps even help us develop a similar ability.
All organisms have the capacity to repair or regenerate tissue damage. None can do it better than salamanders or newts, which can regenerate an entire severed limb.
That feat has amazed and delighted man from the dawn of time and led to endless attempts to understand how it happens – and whether we can control it for our own purposes. An exciting new clue toward that understanding has come from a surprising source: research on the decline of cells, called cellular senescence.
Senescence is the last stage in the life of a cell. Whereas some cells simply break up or wither and die off, others transition into a zombie-like state where they can no longer divide. In this liminal phase, the cell still pumps out many different molecules that can affect its neighbors and cause low grade inflammation. Senescence is associated with many of the declining biological functions that characterize aging, such as inflammation and genomic instability.
Oddly enough, newts are one of the few species that do not accumulate senescent cells as they age, according to research over several years by Maximina Yun. A research group leader at the Center for Regenerative Therapies Dresden and the Max Planck Institute of Molecular and Cell Biology and Genetics, in Dresden, Germany, Yun discovered that senescent cells were induced at some stages of regeneration of the salamander limb, “and then, as the regeneration progresses, they disappeared, they were eliminated by the immune system,” she says. “They were present at particular times and then they disappeared.”
Senescent cells added to the edges of the wound helped the healthy muscle cells to “dedifferentiate,” essentially turning back the developmental clock of those cells into more primitive states.
Previous research on senescence in aging had suggested, logically enough, that applying those cells to the stump of a newly severed salamander limb would slow or even stop its regeneration. But Yun stood that idea on its head. She theorized that senescent cells might also play a role in newt limb regeneration, and she tested it by both adding and removing senescent cells from her animals. It turned out she was right, as the newt limbs grew back faster than normal when more senescent cells were included.
Senescent cells added to the edges of the wound helped the healthy muscle cells to “dedifferentiate,” essentially turning back the developmental clock of those cells into more primitive states, which could then be turned into progenitors, a cell type in between stem cells and specialized cells, needed to regrow the muscle tissue of the missing limb. “We think that this ability to dedifferentiate is intrinsically a big part of why salamanders can regenerate all these very complex structures, which other organisms cannot,” she explains.
Yun sees regeneration as a two part problem. First, the cells must be able to sense that their neighbors from the lost limb are not there anymore. Second, they need to be able to produce the intermediary progenitors for regeneration, , to form what is missing. “Molecularly, that must be encoded like a 3D map,” she says, otherwise the new tissue might grow back as a blob, or liver, or fin instead of a limb.
Wound healing
Another recent study, this time at the Mayo Clinic, provides evidence supporting the role of senescent cells in regeneration. Looking closely at molecules that send information between cells in the wound of a mouse, the researchers found that senescent cells appeared near the start of the healing process and then disappeared as healing progressed. In contrast, persistent senescent cells were the hallmark of a chronic wound that did not heal properly. The function and significance of senescence cells depended on both the timing and the context of their environment.
The paper suggests that senescent cells are not all the same. That has become clearer as researchers have been able to identify protein markers on the surface of some senescent cells. The patterns of these proteins differ for some senescent cells compared to others. In biology, such physical differences suggest functional differences, so it is becoming increasingly likely there are subsets of senescent cells with differing functions that have not yet been identified.
There are disagreements within the research community as to whether newts have acquired their regenerative capacity through a unique evolutionary change, or if other animals, including humans, retain this capacity buried somewhere in their genes.
Scientists initially thought that senescent cells couldn’t play a role in regeneration because they could no longer reproduce, says Anthony Atala, a practicing surgeon and bioengineer who leads the Wake Forest Institute for Regenerative Medicine in North Carolina. But Yun’s study points in the other direction. “What this paper shows clearly is that these cells have the potential to be involved in tissue regeneration [in newts]. The question becomes, will these cells be able to do the same in humans.”
As our knowledge of senescent cells increases, Atala thinks we need to embrace a new analogy to help understand them: humans in retirement. They “have acquired a lot of wisdom throughout their whole life and they can help younger people and mentor them to grow to their full potential. We're seeing the same thing with these cells,” he says. They are no longer putting energy into their own reproduction, but the signaling molecules they secrete “can help other cells around them to regenerate.”
There are disagreements within the research community as to whether newts have acquired their regenerative capacity through a unique evolutionary change, or if other animals, including humans, retain this capacity buried somewhere in their genes. If so, it seems that our genes are unable to express this ability, perhaps as part of a tradeoff in acquiring other traits. It is a fertile area of research.
Dedifferentiation is likely to become an important process in the field of regenerative medicine. One extreme example: a lab has been able to turn back the clock and reprogram adult male skin cells into female eggs, a potential milestone in reproductive health. It will be more difficult to control just how far back one wishes to go in the cell's dedifferentiation – part way or all the way back into a stem cell – and then direct it down a different developmental pathway. Yun is optimistic we can learn these tricks from newts.
Senolytics
A growing field of research is using drugs called senolytics to remove senescent cells and slow or even reverse disease of aging.
“Senolytics are great, but senolytics target different types of senescence,” Yun says. “If senescent cells have positive effects in the context of regeneration, of wound healing, then maybe at the beginning of the regeneration process, you may not want to take them out for a little while.”
“If you look at pretty much all biological systems, too little or too much of something can be bad, you have to be in that central zone” and at the proper time, says Atala. “That's true for proteins, sugars, and the drugs that you take. I think the same thing is true for these cells. Why would they be different?”
Our growing understanding that senescence is not a single thing but a variety of things likely means that effective senolytic drugs will not resemble a single sledge hammer but more a carefully manipulated scalpel where some types of senescent cells are removed while others are added. Combinations and timing could be crucial, meaning the difference between regenerating healthy tissue, a scar, or worse.

