Genomic Data Has a Diversity Problem, But Global Efforts Are Underway to Fix It

Genetic data sets skew too European, threatening to narrow who will benefit from future advances.
Genomics has begun its golden age. Just 20 years ago, sequencing a single genome cost nearly $3 billion and took over a decade. Today, the same feat can be achieved for a few hundred dollars and the better part of a day . Suddenly, the prospect of sequencing not just individuals, but whole populations, has become feasible.
The genetic differences between humans may seem meager, only around 0.1 percent of the genome on average, but this variation can have profound effects on an individual's risk of disease, responsiveness to medication, and even the dosage level that would work best.
Already, initiatives like the U.K.'s 100,000 Genomes Project - now expanding to 1 million genomes - and other similarly massive sequencing projects in Iceland and the U.S., have begun collecting population-scale data in order to capture and study this variation.
The resulting data sets are immensely valuable to researchers and drug developers working to design new 'precision' medicines and diagnostics, and to gain insights that may benefit patients. Yet, because the majority of this data comes from developed countries with well-established scientific and medical infrastructure, the data collected so far is heavily biased towards Western populations with largely European ancestry.
This presents a startling and fast-emerging problem: groups that are under-represented in these datasets are likely to benefit less from the new wave of therapeutics, diagnostics, and insights, simply because they were tailored for the genetic profiles of people with European ancestry.
We may indeed be approaching a golden age of genomics-enabled precision medicine. But if the data bias persists then there is a risk, as with most golden ages throughout history, that the benefits will not be equally accessible to all, and existing inequalities will only be exacerbated.
To remedy the situation, a number of initiatives have sprung up to sequence genomes of under-represented groups, adding them to the datasets and ensuring that they too will benefit from the rapidly unfolding genomic revolution.
Global Gene Corp
The idea behind Global Gene Corp was born eight years ago in Harvard when Sumit Jamuar, co-founder and CEO, met up with his two other co-founders, both experienced geneticists, for a coffee.
"They were discussing the limitless applications of understanding your genetic code," said Jamuar, a business executive from New Delhi.
"And so, being a technology enthusiast type, I was excited and I turned to them and said hey, this is incredible! Could you sequence me and give me some insights? And they actually just turned around and said no, because it's not going to be useful for you - there's not enough reference for what a good Sumit looks like."
What started as a curiosity-driven conversation on the power of genomics ended with a commitment to tackle one of the field's biggest roadblocks - its lack of global representation.
Jamuar set out to begin with India, which has about 20 percent of the world's population, including over 4000 different ethnicities, but contributes less than 2 percent of genomic data, he told Leaps.org.
Eight years later, Global Gene Corp's sequencing initiative is well underway, and is the largest in the history of the Indian subcontinent. The program is being carried out in collaboration with biotech giant Regeneron, with support from the Indian government, local communities, and the Indian healthcare ecosystem. In August 2020, Global Gene Corp's work was recognized through the $1 million 2020 Roddenberry award for organizations that advance the vision of 'Star Trek' creator Gene Roddenberry to better humanity.
This problem has already begun to manifest itself in, for example, much higher levels of genetic misdiagnosis among non-Europeans tested for their risk of certain diseases, such as hypertrophic cardiomyopathy - an inherited disease of the heart muscle.
Global Gene Corp also focuses on developing and implementing AI and machine learning tools to make sense of the deluge of genomic data. These tools are increasingly used by both industry and academia to guide future research by identifying particularly promising or clinically interesting genetic variants. But if the underlying data is skewed European, then the effectiveness of the computational analysis - along with the future advances and avenues of research that emerge from it - will be skewed towards Europeans too.
This problem has already begun to manifest itself in, for example, much higher levels of genetic misdiagnosis among non-Europeans tested for their risk of certain diseases, such as hypertrophic cardiomyopathy - an inherited disease of the heart muscle. Most of the genetic variants used in these tests were identified as being causal for the disease from studies of European genomes. However, many of these variants differ both in their distribution and clinical significance across populations, leading to many patients of non-European ancestry receiving false-positive test results - as their benign genetic variants were misclassified as pathogenic. Had even a small number of genomes from other ethnicities been included in the initial studies, these misdiagnoses could have been avoided.
"Unless we have a data set which is unbiased and representative, we're never going to achieve the success that we want," Jamuar says.
"When Siri was first launched, she could hardly recognize an accent which was not of a certain type, so if I was trying to speak to Siri, I would have to repeat myself multiple times and try to mimic an accent which wasn't my accent so that she could understand it.
"But over time the voice recognition technology improved tremendously because the training data was expanded to include people of very diverse backgrounds and their accents, so the algorithms were trained to be able to pick that up and it dramatically improved the technology. That's the way we have to think about it - without that good-quality diverse data, we will never be able to achieve the full potential of the computational tools."
While mapping India's rich genetic diversity has been the organization's primary focus so far, they plan, in time, to expand their work to other under-represented groups in Asia, the Middle East, Africa, and Latin America.
"As other like-minded people and partners join the mission, it just accelerates the achievement of what we have set out to do, which is to map out and organize the world's genomic diversity so that we can enable high-quality life and longevity benefits for everyone, everywhere," Jamuar says.
Empowering African Genomics
Africa is the birthplace of our species, and today still retains an inordinate amount of total human genetic diversity. Groups that left Africa and went on to populate the rest of the world, some 50 to 100,000 years ago, were likely small in number and only took a fraction of the total genetic diversity with them. This ancient bottleneck means that no other group in the world can match the level of genetic diversity seen in modern African populations.
Despite Africa's central importance in understanding the history and extent of human genetic diversity, the genomics of African populations remains wildly understudied. Addressing this disparity has become a central focus of the H3Africa Consortium, an initiative formally launched in 2012 with support from the African Academy of Sciences, the U.S. National Institutes of Health, and the UK's Wellcome Trust. Today, H3Africa supports over 50 projects across the continent, on an array of different research areas in genetics relevant to the health and heredity of Africans.
"Africa is the cradle of Humankind. So what that really means is that the populations that are currently living in Africa are among some of the oldest populations on the globe, and we know that the longer populations have had to go through evolutionary phases, the more variation there is in the genomes of people who live presently," says Zane Lombard, a principal investigator at H3Africa and Associate Professor of Human Genetics at the University of the Witwatersrand in Johannesburg, South Africa.
"So for that reason, African populations carry a huge amount of genetic variation and diversity, which is pretty much uncaptured. There's still a lot to learn as far as novel variation is concerned by looking at and studying African genomes."
A recent landmark H3Africa study, led by Lombard and published in Nature in October, sequenced the genomes of over 400 African individuals from 50 ethno-linguistic groups - many of which had never been sampled before.
Despite the relatively modest number of individuals sequenced in the study, over three million previously undescribed genetic variants were found, and complex patterns of ancestral migration were uncovered.
"In some of these ethno-linguistic groups they don't have a word for DNA, so we've had to really think about how to make sure that we communicate the purposes of different studies to participants so that you have true informed consent," says Lombard.
"The objective," she explained, "was to try and fill some of the gaps for many of these populations for which we didn't have any whole genome sequences or any genetic variation data...because if we're thinking about the future of precision medicine, if the patient is a member of a specific group where we don't know a lot about the genomic variation that exists in that group, it makes it really difficult to start thinking about clinical interpretation of their data."
From H3Africa's conception, the consortium's goal has not only been to better represent Africa's staggering genetic diversity in genomic data sets, but also to build Africa's domestic genomics capabilities and empower a new generation of African researchers. By doing so, the hope is that Africans will be able to set their own genomics agenda, and leapfrog to new and better ways of doing the work.
"The training that has happened on the continent and the number of new scientists, new students, and fellows that have come through the process and are now enabled to start their own research groups, to grow their own research in their countries, to be a spokesperson for genomics research in their countries, and to build that political will to do these larger types of sequencing initiatives - that is really a significant outcome from H3Africa as well. Over and above all the science that's coming out," Lombard says.
"What has been created through H3Africa is just this locus of researchers and scientists and bioethicists who have the same goal at heart - to work towards adjusting the data bias and making sure that all global populations are represented in genomics."
Thanks to safety cautions from the COVID-19 pandemic, a strain of influenza has been completely eliminated.
If you were one of the millions who masked up, washed your hands thoroughly and socially distanced, pat yourself on the back—you may have helped change the course of human history.
Scientists say that thanks to these safety precautions, which were introduced in early 2020 as a way to stop transmission of the novel COVID-19 virus, a strain of influenza has been completely eliminated. This marks the first time in human history that a virus has been wiped out through non-pharmaceutical interventions, such as vaccines.
The flu shot, explained
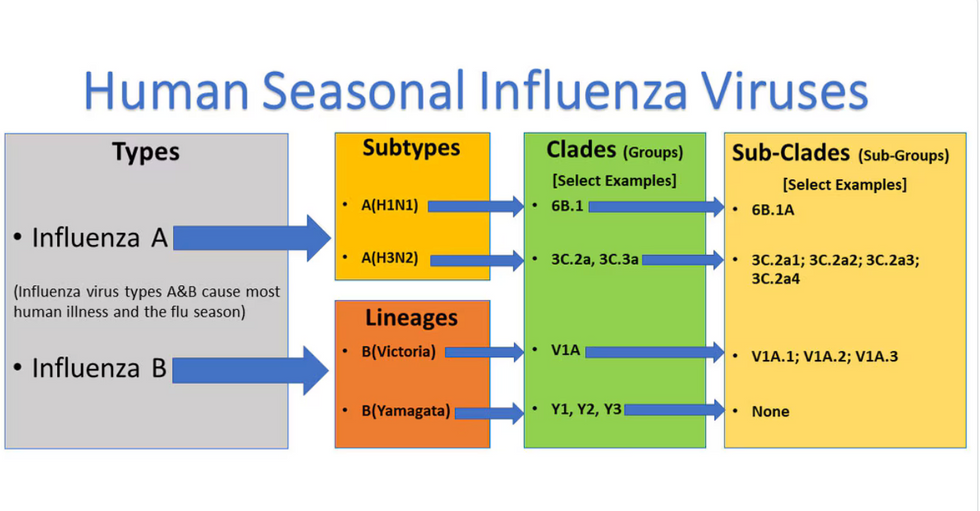
Influenza viruses type A and B are responsible for the majority of human illnesses and the flu season.
Centers for Disease Control
For more than a decade, flu shots have protected against two types of the influenza virus–type A and type B. While there are four different strains of influenza in existence (A, B, C, and D), only strains A, B, and C are capable of infecting humans, and only A and B cause pandemics. In other words, if you catch the flu during flu season, you’re most likely sick with flu type A or B.
Flu vaccines contain inactivated—or dead—influenza virus. These inactivated viruses can’t cause sickness in humans, but when administered as part of a vaccine, they teach a person’s immune system to recognize and kill those viruses when they’re encountered in the wild.
Each spring, a panel of experts gives a recommendation to the US Food and Drug Administration on which strains of each flu type to include in that year’s flu vaccine, depending on what surveillance data says is circulating and what they believe is likely to cause the most illness during the upcoming flu season. For the past decade, Americans have had access to vaccines that provide protection against two strains of influenza A and two lineages of influenza B, known as the Victoria lineage and the Yamagata lineage. But this year, the seasonal flu shot won’t include the Yamagata strain, because the Yamagata strain is no longer circulating among humans.
How Yamagata Disappeared

Flu surveillance data from the Global Initiative on Sharing All Influenza Data (GISAID) shows that the Yamagata lineage of flu type B has not been sequenced since April 2020.
Nature
Experts believe that the Yamagata lineage had already been in decline before the pandemic hit, likely because the strain was naturally less capable of infecting large numbers of people compared to the other strains. When the COVID-19 pandemic hit, the resulting safety precautions such as social distancing, isolating, hand-washing, and masking were enough to drive the virus into extinction completely.
Because the strain hasn’t been circulating since 2020, the FDA elected to remove the Yamagata strain from the seasonal flu vaccine. This will mark the first time since 2012 that the annual flu shot will be trivalent (three-component) rather than quadrivalent (four-component).
Should I still get the flu shot?
The flu shot will protect against fewer strains this year—but that doesn’t mean we should skip it. Influenza places a substantial health burden on the United States every year, responsible for hundreds of thousands of hospitalizations and tens of thousands of deaths. The flu shot has been shown to prevent millions of illnesses each year (more than six million during the 2022-2023 season). And while it’s still possible to catch the flu after getting the flu shot, studies show that people are far less likely to be hospitalized or die when they’re vaccinated.
Another unexpected benefit of dropping the Yamagata strain from the seasonal vaccine? This will possibly make production of the flu vaccine faster, and enable manufacturers to make more vaccines, helping countries who have a flu vaccine shortage and potentially saving millions more lives.
After his grandmother’s dementia diagnosis, one man invented a snack to keep her healthy and hydrated.
Founder Lewis Hornby and his grandmother Pat, sampling Jelly Drops—an edible gummy containing water and life-saving electrolytes.
On a visit to his grandmother’s nursing home in 2016, college student Lewis Hornby made a shocking discovery: Dehydration is a common (and dangerous) problem among seniors—especially those that are diagnosed with dementia.
Hornby’s grandmother, Pat, had always had difficulty keeping up her water intake as she got older, a common issue with seniors. As we age, our body composition changes, and we naturally hold less water than younger adults or children, so it’s easier to become dehydrated quickly if those fluids aren’t replenished. What’s more, our thirst signals diminish naturally as we age as well—meaning our body is not as good as it once was in letting us know that we need to rehydrate. This often creates a perfect storm that commonly leads to dehydration. In Pat’s case, her dehydration was so severe she nearly died.
When Lewis Hornby visited his grandmother at her nursing home afterward, he learned that dehydration especially affects people with dementia, as they often don’t feel thirst cues at all, or may not recognize how to use cups correctly. But while dementia patients often don’t remember to drink water, it seemed to Hornby that they had less problem remembering to eat, particularly candy.

Hornby wanted to create a solution for elderly people who struggled keeping their fluid intake up. He spent the next eighteen months researching and designing a solution and securing funding for his project. In 2019, Hornby won a sizable grant from the Alzheimer’s Society, a UK-based care and research charity for people with dementia and their caregivers. Together, through the charity’s Accelerator Program, they created a bite-sized, sugar-free, edible jelly drop that looked and tasted like candy. The candy, called Jelly Drops, contained 95% water and electrolytes—important minerals that are often lost during dehydration. The final product launched in 2020—and was an immediate success. The drops were able to provide extra hydration to the elderly, as well as help keep dementia patients safe, since dehydration commonly leads to confusion, hospitalization, and sometimes even death.
Not only did Jelly Drops quickly become a favorite snack among dementia patients in the UK, but they were able to provide an additional boost of hydration to hospital workers during the pandemic. In NHS coronavirus hospital wards, patients infected with the virus were regularly given Jelly Drops to keep their fluid levels normal—and staff members snacked on them as well, since long shifts and personal protective equipment (PPE) they were required to wear often left them feeling parched.
In April 2022, Jelly Drops launched in the United States. The company continues to donate 1% of its profits to help fund Alzheimer’s research.

