Genetically Sequencing Healthy Babies Yielded Surprising Results

A newborn and mother in the hospital - first touch. (© martin81/Shutterstock)
Today in Melrose, Massachusetts, Cora Stetson is the picture of good health, a bubbly precocious 2-year-old. But Cora has two separate mutations in the gene that produces a critical enzyme called biotinidase and her body produces only 40 percent of the normal levels of that enzyme.
In the last few years, the dream of predicting and preventing diseases through genomics, starting in childhood, is finally within reach.
That's enough to pass conventional newborn (heelstick) screening, but may not be enough for normal brain development, putting baby Cora at risk for seizures and cognitive impairment. But thanks to an experimental study in which Cora's DNA was sequenced after birth, this condition was discovered and she is being treated with a safe and inexpensive vitamin supplement.
Stories like these are beginning to emerge from the BabySeq Project, the first clinical trial in the world to systematically sequence healthy newborn infants. This trial was led by my research group with funding from the National Institutes of Health. While still controversial, it is pointing the way to a future in which adults, or even newborns, can receive comprehensive genetic analysis in order to determine their risk of future disease and enable opportunities to prevent them.
Some believe that medicine is still not ready for genomic population screening, but others feel it is long overdue. After all, the sequencing of the Human Genome Project was completed in 2003, and with this milestone, it became feasible to sequence and interpret the genome of any human being. The costs have come down dramatically since then; an entire human genome can now be sequenced for about $800, although the costs of bioinformatic and medical interpretation can add another $200 to $2000 more, depending upon the number of genes interrogated and the sophistication of the interpretive effort.

Two-year-old Cora Stetson, whose DNA sequencing after birth identified a potentially dangerous genetic mutation in time for her to receive preventive treatment.
(Photo courtesy of Robert Green)
The ability to sequence the human genome yielded extraordinary benefits in scientific discovery, disease diagnosis, and targeted cancer treatment. But the ability of genomes to detect health risks in advance, to actually predict the medical future of an individual, has been mired in controversy and slow to manifest. In particular, the oft-cited vision that healthy infants could be genetically tested at birth in order to predict and prevent the diseases they would encounter, has proven to be far tougher to implement than anyone anticipated.
But in the last few years, the dream of predicting and preventing diseases through genomics, starting in childhood, is finally within reach. Why did it take so long? And what remains to be done?
Great Expectations
Part of the problem was the unrealistic expectations that had been building for years in advance of the genomic science itself. For example, the 1997 film Gattaca portrayed a near future in which the lifetime risk of disease was readily predicted the moment an infant is born. In the fanfare that accompanied the completion of the Human Genome Project, the notion of predicting and preventing future disease in an individual became a powerful meme that was used to inspire investment and public support for genomic research long before the tools were in place to make it happen.
Another part of the problem was the success of state-mandated newborn screening programs that began in the 1960's with biochemical tests of the "heel-stick" for babies with metabolic disorders. These programs have worked beautifully, costing only a few dollars per baby and saving thousands of infants from death and severe cognitive impairment. It seemed only logical that a new technology like genome sequencing would add power and promise to such programs. But instead of embracing the notion of newborn sequencing, newborn screening laboratories have thus far rejected the entire idea as too expensive, too ambiguous, and too threatening to the comfortable constituency that they had built within the public health framework.
"What can you find when you look as deeply as possible into the medical genomes of healthy individuals?"
Creating the Evidence Base for Preventive Genomics
Despite a number of obstacles, there are researchers who are exploring how to achieve the original vision of genomic testing as a tool for disease prediction and prevention. For example, in our NIH-funded MedSeq Project, we were the first to ask the question: "What can you find when you look as deeply as possible into the medical genomes of healthy individuals?"
Most people do not understand that genetic information comes in four separate categories: 1) dominant mutations putting the individual at risk for rare conditions like familial forms of heart disease or cancer, (2) recessive mutations putting the individual's children at risk for rare conditions like cystic fibrosis or PKU, (3) variants across the genome that can be tallied to construct polygenic risk scores for common conditions like heart disease or type 2 diabetes, and (4) variants that can influence drug metabolism or predict drug side effects such as the muscle pain that occasionally occurs with statin use.
The technological and analytical challenges of our study were formidable, because we decided to systematically interrogate over 5000 disease-associated genes and report results in all four categories of genetic information directly to the primary care physicians for each of our volunteers. We enrolled 200 adults and found that everyone who was sequenced had medically relevant polygenic and pharmacogenomic results, over 90 percent carried recessive mutations that could have been important to reproduction, and an extraordinary 14.5 percent carried dominant mutations for rare genetic conditions.
A few years later we launched the BabySeq Project. In this study, we restricted the number of genes to include only those with child/adolescent onset that could benefit medically from early warning, and even so, we found 9.4 percent carried dominant mutations for rare conditions.
At first, our interpretation around the high proportion of apparently healthy individuals with dominant mutations for rare genetic conditions was simple – that these conditions had lower "penetrance" than anticipated; in other words, only a small proportion of those who carried the dominant mutation would get the disease. If this interpretation were to hold, then genetic risk information might be far less useful than we had hoped.
Suddenly the information available in the genome of even an apparently healthy individual is looking more robust, and the prospect of preventive genomics is looking feasible.
But then we circled back with each adult or infant in order to examine and test them for any possible features of the rare disease in question. When we did this, we were surprised to see that in over a quarter of those carrying such mutations, there were already subtle signs of the disease in question that had not even been suspected! Now our interpretation was different. We now believe that genetic risk may be responsible for subclinical disease in a much higher proportion of people than has ever been suspected!
Meanwhile, colleagues of ours have been demonstrating that detailed analysis of polygenic risk scores can identify individuals at high risk for common conditions like heart disease. So adding up the medically relevant results in any given genome, we start to see that you can learn your risks for a rare monogenic condition, a common polygenic condition, a bad effect from a drug you might take in the future, or for having a child with a devastating recessive condition. Suddenly the information available in the genome of even an apparently healthy individual is looking more robust, and the prospect of preventive genomics is looking feasible.
Preventive Genomics Arrives in Clinical Medicine
There is still considerable evidence to gather before we can recommend genomic screening for the entire population. For example, it is important to make sure that families who learn about such risks do not suffer harms or waste resources from excessive medical attention. And many doctors don't yet have guidance on how to use such information with their patients. But our research is convincing many people that preventive genomics is coming and that it will save lives.
In fact, we recently launched a Preventive Genomics Clinic at Brigham and Women's Hospital where information-seeking adults can obtain predictive genomic testing with the highest quality interpretation and medical context, and be coached over time in light of their disease risks toward a healthier outcome. Insurance doesn't yet cover such testing, so patients must pay out of pocket for now, but they can choose from a menu of genetic screening tests, all of which are more comprehensive than consumer-facing products. Genetic counseling is available but optional. So far, this service is for adults only, but sequencing for children will surely follow soon.
As the costs of sequencing and other Omics technologies continue to decline, we will see both responsible and irresponsible marketing of genetic testing, and we will need to guard against unscientific claims. But at the same time, we must be far more imaginative and fast moving in mainstream medicine than we have been to date in order to claim the emerging benefits of preventive genomics where it is now clear that suffering can be averted, and lives can be saved. The future has arrived if we are bold enough to grasp it.
Funding and Disclosures:
Dr. Green's research is supported by the National Institutes of Health, the Department of Defense and through donations to The Franca Sozzani Fund for Preventive Genomics. Dr. Green receives compensation for advising the following companies: AIA, Applied Therapeutics, Helix, Ohana, OptraHealth, Prudential, Verily and Veritas; and is co-founder and advisor to Genome Medical, Inc, a technology and services company providing genetics expertise to patients, providers, employers and care systems.
A robot server, controlled remotely by a disabled worker, delivers drinks to patrons at the DAWN cafe in Tokyo.
A sleek, four-foot tall white robot glides across a cafe storefront in Tokyo’s Nihonbashi district, holding a two-tiered serving tray full of tea sandwiches and pastries. The cafe’s patrons smile and say thanks as they take the tray—but it’s not the robot they’re thanking. Instead, the patrons are talking to the person controlling the robot—a restaurant employee who operates the avatar from the comfort of their home.
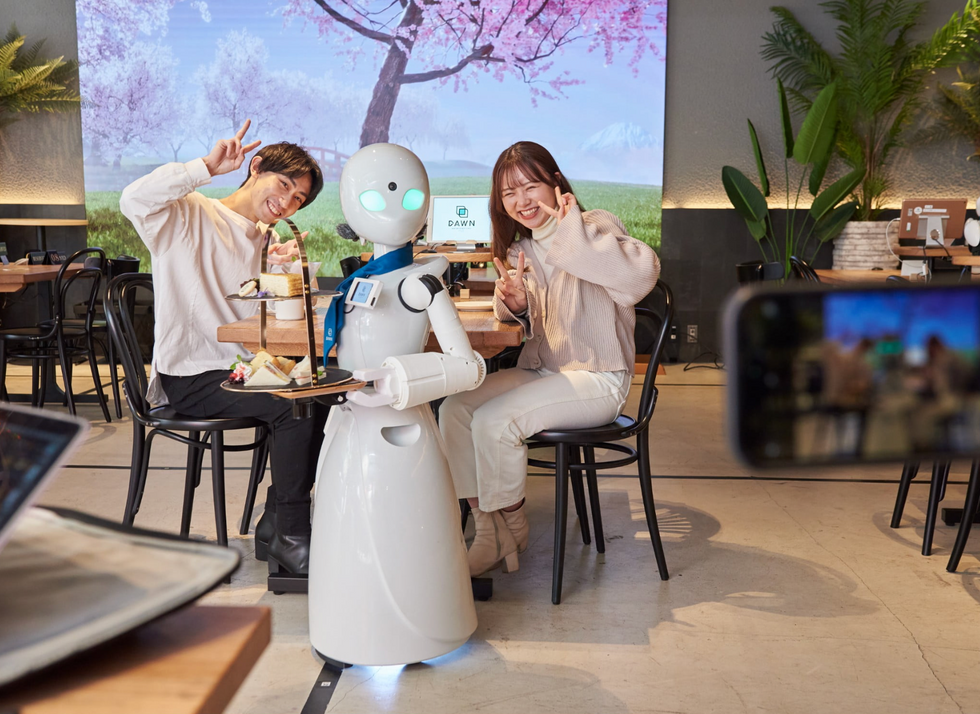
It’s a typical scene at DAWN, short for Diverse Avatar Working Network—a cafe that launched in Tokyo six years ago as an experimental pop-up and quickly became an overnight success. Today, the cafe is a permanent fixture in Nihonbashi, staffing roughly 60 remote workers who control the robots remotely and communicate to customers via a built-in microphone.
More than just a creative idea, however, DAWN is being hailed as a life-changing opportunity. The workers who control the robots remotely (known as “pilots”) all have disabilities that limit their ability to move around freely and travel outside their homes. Worldwide, an estimated 16 percent of the global population lives with a significant disability—and according to the World Health Organization, these disabilities give rise to other problems, such as exclusion from education, unemployment, and poverty.
These are all problems that Kentaro Yoshifuji, founder and CEO of Ory Laboratory, which supplies the robot servers at DAWN, is looking to correct. Yoshifuji, who was bedridden for several years in high school due to an undisclosed health problem, launched the company to help enable people who are house-bound or bedridden to more fully participate in society, as well as end the loneliness, isolation, and feelings of worthlessness that can sometimes go hand-in-hand with being disabled.
“It’s heartbreaking to think that [people with disabilities] feel they are a burden to society, or that they fear their families suffer by caring for them,” said Yoshifuji in an interview in 2020. “We are dedicating ourselves to providing workable, technology-based solutions. That is our purpose.”
Shota, Kuwahara, a DAWN employee with muscular dystrophy, agrees. "There are many difficulties in my daily life, but I believe my life has a purpose and is not being wasted," he says. "Being useful, able to help other people, even feeling needed by others, is so motivational."
A woman receives a mammogram, which can detect the presence of tumors in a patient's breast.
When a patient is diagnosed with early-stage breast cancer, having surgery to remove the tumor is considered the standard of care. But what happens when a patient can’t have surgery?
Whether it’s due to high blood pressure, advanced age, heart issues, or other reasons, some breast cancer patients don’t qualify for a lumpectomy—one of the most common treatment options for early-stage breast cancer. A lumpectomy surgically removes the tumor while keeping the patient’s breast intact, while a mastectomy removes the entire breast and nearby lymph nodes.
Fortunately, a new technique called cryoablation is now available for breast cancer patients who either aren’t candidates for surgery or don’t feel comfortable undergoing a surgical procedure. With cryoablation, doctors use an ultrasound or CT scan to locate any tumors inside the patient’s breast. They then insert small, needle-like probes into the patient's breast which create an “ice ball” that surrounds the tumor and kills the cancer cells.
Cryoablation has been used for decades to treat cancers of the kidneys and liver—but only in the past few years have doctors been able to use the procedure to treat breast cancer patients. And while clinical trials have shown that cryoablation works for tumors smaller than 1.5 centimeters, a recent clinical trial at Memorial Sloan Kettering Cancer Center in New York has shown that it can work for larger tumors, too.
In this study, doctors performed cryoablation on patients whose tumors were, on average, 2.5 centimeters. The cryoablation procedure lasted for about 30 minutes, and patients were able to go home on the same day following treatment. Doctors then followed up with the patients after 16 months. In the follow-up, doctors found the recurrence rate for tumors after using cryoablation was only 10 percent.
For patients who don’t qualify for surgery, radiation and hormonal therapy is typically used to treat tumors. However, said Yolanda Brice, M.D., an interventional radiologist at Memorial Sloan Kettering Cancer Center, “when treated with only radiation and hormonal therapy, the tumors will eventually return.” Cryotherapy, Brice said, could be a more effective way to treat cancer for patients who can’t have surgery.
“The fact that we only saw a 10 percent recurrence rate in our study is incredibly promising,” she said.