Genetically Sequencing Healthy Babies Yielded Surprising Results

A newborn and mother in the hospital - first touch. (© martin81/Shutterstock)
Today in Melrose, Massachusetts, Cora Stetson is the picture of good health, a bubbly precocious 2-year-old. But Cora has two separate mutations in the gene that produces a critical enzyme called biotinidase and her body produces only 40 percent of the normal levels of that enzyme.
In the last few years, the dream of predicting and preventing diseases through genomics, starting in childhood, is finally within reach.
That's enough to pass conventional newborn (heelstick) screening, but may not be enough for normal brain development, putting baby Cora at risk for seizures and cognitive impairment. But thanks to an experimental study in which Cora's DNA was sequenced after birth, this condition was discovered and she is being treated with a safe and inexpensive vitamin supplement.
Stories like these are beginning to emerge from the BabySeq Project, the first clinical trial in the world to systematically sequence healthy newborn infants. This trial was led by my research group with funding from the National Institutes of Health. While still controversial, it is pointing the way to a future in which adults, or even newborns, can receive comprehensive genetic analysis in order to determine their risk of future disease and enable opportunities to prevent them.
Some believe that medicine is still not ready for genomic population screening, but others feel it is long overdue. After all, the sequencing of the Human Genome Project was completed in 2003, and with this milestone, it became feasible to sequence and interpret the genome of any human being. The costs have come down dramatically since then; an entire human genome can now be sequenced for about $800, although the costs of bioinformatic and medical interpretation can add another $200 to $2000 more, depending upon the number of genes interrogated and the sophistication of the interpretive effort.

Two-year-old Cora Stetson, whose DNA sequencing after birth identified a potentially dangerous genetic mutation in time for her to receive preventive treatment.
(Photo courtesy of Robert Green)
The ability to sequence the human genome yielded extraordinary benefits in scientific discovery, disease diagnosis, and targeted cancer treatment. But the ability of genomes to detect health risks in advance, to actually predict the medical future of an individual, has been mired in controversy and slow to manifest. In particular, the oft-cited vision that healthy infants could be genetically tested at birth in order to predict and prevent the diseases they would encounter, has proven to be far tougher to implement than anyone anticipated.
But in the last few years, the dream of predicting and preventing diseases through genomics, starting in childhood, is finally within reach. Why did it take so long? And what remains to be done?
Great Expectations
Part of the problem was the unrealistic expectations that had been building for years in advance of the genomic science itself. For example, the 1997 film Gattaca portrayed a near future in which the lifetime risk of disease was readily predicted the moment an infant is born. In the fanfare that accompanied the completion of the Human Genome Project, the notion of predicting and preventing future disease in an individual became a powerful meme that was used to inspire investment and public support for genomic research long before the tools were in place to make it happen.
Another part of the problem was the success of state-mandated newborn screening programs that began in the 1960's with biochemical tests of the "heel-stick" for babies with metabolic disorders. These programs have worked beautifully, costing only a few dollars per baby and saving thousands of infants from death and severe cognitive impairment. It seemed only logical that a new technology like genome sequencing would add power and promise to such programs. But instead of embracing the notion of newborn sequencing, newborn screening laboratories have thus far rejected the entire idea as too expensive, too ambiguous, and too threatening to the comfortable constituency that they had built within the public health framework.
"What can you find when you look as deeply as possible into the medical genomes of healthy individuals?"
Creating the Evidence Base for Preventive Genomics
Despite a number of obstacles, there are researchers who are exploring how to achieve the original vision of genomic testing as a tool for disease prediction and prevention. For example, in our NIH-funded MedSeq Project, we were the first to ask the question: "What can you find when you look as deeply as possible into the medical genomes of healthy individuals?"
Most people do not understand that genetic information comes in four separate categories: 1) dominant mutations putting the individual at risk for rare conditions like familial forms of heart disease or cancer, (2) recessive mutations putting the individual's children at risk for rare conditions like cystic fibrosis or PKU, (3) variants across the genome that can be tallied to construct polygenic risk scores for common conditions like heart disease or type 2 diabetes, and (4) variants that can influence drug metabolism or predict drug side effects such as the muscle pain that occasionally occurs with statin use.
The technological and analytical challenges of our study were formidable, because we decided to systematically interrogate over 5000 disease-associated genes and report results in all four categories of genetic information directly to the primary care physicians for each of our volunteers. We enrolled 200 adults and found that everyone who was sequenced had medically relevant polygenic and pharmacogenomic results, over 90 percent carried recessive mutations that could have been important to reproduction, and an extraordinary 14.5 percent carried dominant mutations for rare genetic conditions.
A few years later we launched the BabySeq Project. In this study, we restricted the number of genes to include only those with child/adolescent onset that could benefit medically from early warning, and even so, we found 9.4 percent carried dominant mutations for rare conditions.
At first, our interpretation around the high proportion of apparently healthy individuals with dominant mutations for rare genetic conditions was simple – that these conditions had lower "penetrance" than anticipated; in other words, only a small proportion of those who carried the dominant mutation would get the disease. If this interpretation were to hold, then genetic risk information might be far less useful than we had hoped.
Suddenly the information available in the genome of even an apparently healthy individual is looking more robust, and the prospect of preventive genomics is looking feasible.
But then we circled back with each adult or infant in order to examine and test them for any possible features of the rare disease in question. When we did this, we were surprised to see that in over a quarter of those carrying such mutations, there were already subtle signs of the disease in question that had not even been suspected! Now our interpretation was different. We now believe that genetic risk may be responsible for subclinical disease in a much higher proportion of people than has ever been suspected!
Meanwhile, colleagues of ours have been demonstrating that detailed analysis of polygenic risk scores can identify individuals at high risk for common conditions like heart disease. So adding up the medically relevant results in any given genome, we start to see that you can learn your risks for a rare monogenic condition, a common polygenic condition, a bad effect from a drug you might take in the future, or for having a child with a devastating recessive condition. Suddenly the information available in the genome of even an apparently healthy individual is looking more robust, and the prospect of preventive genomics is looking feasible.
Preventive Genomics Arrives in Clinical Medicine
There is still considerable evidence to gather before we can recommend genomic screening for the entire population. For example, it is important to make sure that families who learn about such risks do not suffer harms or waste resources from excessive medical attention. And many doctors don't yet have guidance on how to use such information with their patients. But our research is convincing many people that preventive genomics is coming and that it will save lives.
In fact, we recently launched a Preventive Genomics Clinic at Brigham and Women's Hospital where information-seeking adults can obtain predictive genomic testing with the highest quality interpretation and medical context, and be coached over time in light of their disease risks toward a healthier outcome. Insurance doesn't yet cover such testing, so patients must pay out of pocket for now, but they can choose from a menu of genetic screening tests, all of which are more comprehensive than consumer-facing products. Genetic counseling is available but optional. So far, this service is for adults only, but sequencing for children will surely follow soon.
As the costs of sequencing and other Omics technologies continue to decline, we will see both responsible and irresponsible marketing of genetic testing, and we will need to guard against unscientific claims. But at the same time, we must be far more imaginative and fast moving in mainstream medicine than we have been to date in order to claim the emerging benefits of preventive genomics where it is now clear that suffering can be averted, and lives can be saved. The future has arrived if we are bold enough to grasp it.
Funding and Disclosures:
Dr. Green's research is supported by the National Institutes of Health, the Department of Defense and through donations to The Franca Sozzani Fund for Preventive Genomics. Dr. Green receives compensation for advising the following companies: AIA, Applied Therapeutics, Helix, Ohana, OptraHealth, Prudential, Verily and Veritas; and is co-founder and advisor to Genome Medical, Inc, a technology and services company providing genetics expertise to patients, providers, employers and care systems.
Kirstie Ennis, an Afghanistan veteran who survived a helicopter crash but lost a limb, pictured in May 2021 at Two Rivers Park in Colorado.
In June 2012, Kirstie Ennis was six months into her second deployment to Afghanistan and recently promoted to sergeant. The helicopter gunner and seven others were three hours into a routine mission of combat resupplies and troop transport when their CH-53D helicopter went down hard.
Miraculously, all eight people onboard survived, but Ennis' injuries were many and severe. She had a torn rotator cuff, torn labrum, crushed cervical discs, facial fractures, deep lacerations and traumatic brain injury. Despite a severely fractured ankle, doctors managed to save her foot, for a while at least.
In November 2015, after three years of constant pain and too many surgeries to count, Ennis relented. She elected to undergo a lower leg amputation but only after she completed the 1,000-mile, 72-day Walking with the Wounded journey across the UK.
On Veteran's Day of that year, on the other side of the country, orthopedic surgeon Cato Laurencin announced a moonshot challenge he was setting out to achieve on behalf of wounded warriors like Ennis: the Hartford Engineering A Limb (HEAL) Project.
Laurencin, who is a University of Connecticut professor of chemical, materials and biomedical engineering, teamed up with experts in tissue bioengineering and regenerative medicine from Harvard, Columbia, UC Irvine and SASTRA University in India. Laurencin and his colleagues at the Connecticut Convergence Institute for Translation in Regenerative Engineering made a bold commitment to regenerate an entire limb within 15 years – by the year 2030.

Dr. Cato Laurencin pictured in his office at UConn.
Photo Credit: UConn
Regenerative Engineering -- A Whole New Field
Limb regeneration in humans has been a medical and scientific fascination for decades, with little to show for the effort. However, Laurencin believes that if we are to reach the next level of 21st century medical advances, this puzzle must be solved.
An estimated 185,000 people undergo upper or lower limb amputation every year. Despite the significant advances in electromechanical prosthetics, these individuals still lack the ability to perform complex functions such as sensation for tactile input, normal gait and movement feedback. As far as Laurencin is concerned, the only clinical answer that makes sense is to regenerate a whole functional limb.
Laurencin feels other regeneration efforts were hampered by their siloed research methods with chemists, surgeons, engineers all working separately. Success, he argues, requires a paradigm shift to a trans-disciplinary approach that brings together cutting-edge technologies from disparate fields such as biology, material sciences, physical, chemical and engineering sciences.
As the only surgeon ever inducted into the academies of Science, Medicine and Innovation, Laurencin is uniquely suited for the challenge. He is regarded as the founder of Regenerative Engineering, defined as the convergence of advanced materials sciences, stem cell sciences, physics, developmental biology and clinical translation for the regeneration of complex tissues and organ systems.
But none of this is achievable without early clinician participation across scientific fields to develop new technologies and a deeper understanding of how to harness the body's innate regenerative capabilities. "When I perform a surgical procedure or something is torn or needs to be repaired, I count on the body being involved in regenerating tissue," he says. "So, understanding how the body works to regenerate itself and harnessing that ability is an important factor for the regeneration process."
The Birth of the Vision
Laurencin's passion for regeneration began when he was a sports medicine fellow at Cornell University Medical Center in the early 1990s. There he saw a significant number of injuries to the anterior cruciate ligament (ACL), the major ligament that stabilizes the knee. He believed he could develop a better way to address those injuries using biomaterials to regenerate the ligament. He sketched out a preliminary drawing on a napkin one night over dinner. He has spent the next 30 years regenerating tissues, including the patented L-C ligament.
As chair of Orthopaedic Surgery at the University of Virginia during the peak of the wars in Iraq and Afghanistan, Laurencin treated military personnel who survived because of improved helmets, body armor and battlefield medicine but were left with more devastating injuries, including traumatic brain injuries and limb loss.
"I was so honored to care for them and I so admired their steadfast courage that I became determined to do something big for them," says Laurencin.
When he tells people about his plans to regrow a limb, he gets a lot of eye rolls, which he finds amusing but not discouraging. Growing bone cells was relatively new when he was first focused on regenerating bone in 1987 at MIT; in 2007 he was well on his way to regenerating ligaments at UVA when many still doubted that ligaments could even be reconstructed. He and his team have already regenerated torn rotator cuff tendons and ACL ligaments using a nano-textured fabric seeded with stem cells.
Even as a finalist for the $4 million NIH Pioneer Award for high-risk/high-reward research, he faced a skeptical scientific audience in 2014. "They said, 'Well what do you plan to do?' I said 'I plan to regenerate a whole limb in people.' There was a lot of incredulousness. They stared at me and asked a lot of questions. About three days later, I received probably the best score I've ever gotten on an NIH grant."
In the Thick of the Science
Humans are born with regenerative abilities--two-year-olds have regrown fingertips--but lose that ability with age. Salamanders are the only vertebrates that can regenerate lost body parts as adults; axolotl, the rare Mexican salamander, can grow extra limbs.
The axolotl is important as a model organism because it is a four-footed vertebrate with a similar body plan to humans. Mapping the axolotl genome in 2018 enhanced scientists' genetic understanding of their evolution, development, and regeneration. Being easy to breed in captivity allowed the HEAL team to closely study these amphibians and discover a new cell type they believe may shed light on how to mimic the process in humans.
"Whenever limb regeneration takes place in the salamander, there is a huge amount of something called heparan sulfate around that area," explains Laurencin. "We thought, 'What if this heparan sulfate is the key ingredient to allowing regeneration to take place?' We found these groups of cells that were interspersed in tissues during the time of regeneration that seemed to have connections to each other that expressed this heparan sulfate."
Called GRID (Groups that are Regenerative, Interspersed and Dendritic), these cells were also recently discovered in mice. While GRID cells don't regenerate as well in mice as in salamanders, finding them in mammals was significant.
"If they're found in mice. we might be able to find these in humans in some form," Laurencin says. "We think maybe it will help us figure out regeneration or we can create cells that mimic what grid cells do and create an artificial grid cell."
What Comes Next?
Laurencin and his team have individually engineered and made every single tissue in the lower limb, including bone, cartilage, ligament, skin, nerve, blood vessels. Regenerating joints and joint tissue is the next big mile marker, which Laurencin sees as essential to regenerating a limb that functions and performs in the way he envisions.
"Using stem cells and amnion tissue, we can regenerate joints that are damaged, and have severe arthritis," he says. "We're making progress on all fronts, and making discoveries we believe are going to be helping people along the way."
That focus and advancement is vital to Ennis. After laboring over the decision to have her leg amputated below the knee, she contracted MRSA two weeks post-surgery. In less than a month, she went from a below-the-knee-amputee to a through-the-knee amputee to an above-the-knee amputee.
"A below-the-knee amputation is night-and-day from above-the-knee," she said. "You have to relearn everything. You're basically a toddler."
Kirstie Ennis pictured in July 2020.
Photo Credit: Ennis' Instagram
The clock is ticking on the timeline Laurencin set for himself. Nine years might seem like forever if you're doing time but it might appear fleeting when you're trying to create something that's never been done before. But Laurencin isn't worried. He's convinced time is on his side.
"Every week, I receive an email or a call from someone, maybe a mother whose child has lost a finger or I'm in communication with a disabled American veteran who wants to know how the progress is going. That energizes me to continue to work hard to try to create these sorts of solutions because we're talking about people and their lives."
He devotes about 60 hours a week to the project and the roughly 100 students, faculty and staff who make up the HEAL team at the Convergence Institute seem acutely aware of what's at stake and appear equally dedicated.
"We're in the thick of the science in terms of making this happen," says Laurencin. "We've moved from making the impossible possible to making the possible a reality. That's what science is all about."
7 Reasons Why We Should Not Need Boosters for COVID-19
A top infectious disease physician explains why immunity derived from natural infection and the vaccines should be long-lasting.
There are at least 7 reasons why immunity after vaccination or infection with COVID-19 should likely be long-lived. If durable, I do not think boosters will be necessary in the future, despite CEOs of pharmaceutical companies (who stand to profit from boosters) messaging that they may and readying such boosters. To explain these reasons, let's orient ourselves to the main components of the immune system.
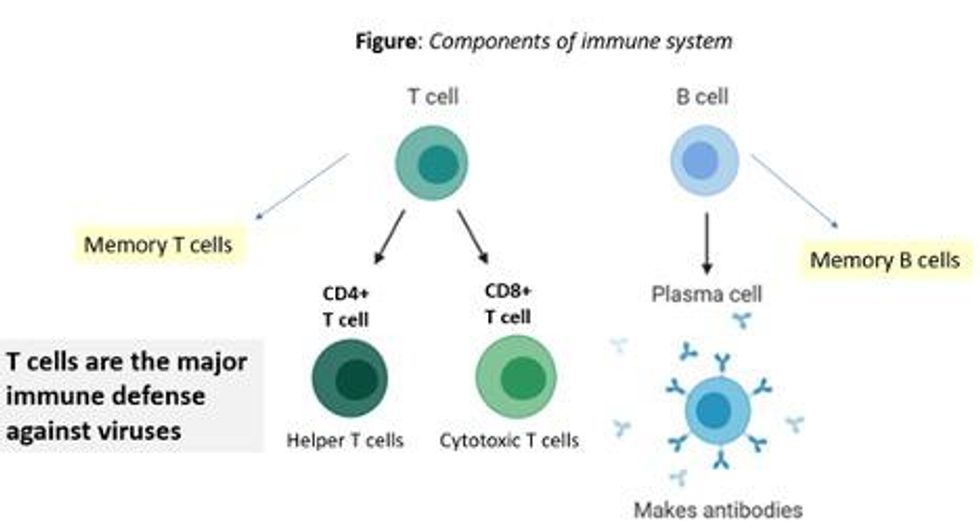
There are two major arms of the immune system: B cells (which produce antibodies) and T cells (which are formed specifically to attack and kill pathogens). T cells are divided into two types, CD4 cells ("helper" T cells) and CD8 cells ("cytotoxic" T cells).
Each arm, once stimulated by infection or vaccine, should hopefully make "memory" banks. So if the body sees the pathogen in the future, these defenses should come roaring back to attack the virus and protect you from getting sick. Plenty of research in COVID-19 indicates a likely long-lasting response to the vaccine or infection. Here are seven of the most compelling reasons:
REASON 1: Memory B Cells Are Produced By Vaccines and Natural Infection
In one study, 12 volunteers who had never had Covid-19--and were fully vaccinated with two Pfizer/BioNTech shots-- underwent biopsies of their lymph nodes. This is where memory B cells are stored in places called "germinal centers". The biopsies were performed three, four, six, and seven weeks after the first mRNA vaccine shot, and were stained to reveal that germinal center memory B cells in the lymph nodes increased in concentration over time.
Natural infection also generates memory B cells. Even after antibody levels wane over time, strong memory B cells were detected in the blood of individuals six and eight months after infection in different studies. Indeed, the half-lives of the memory B cells seen in the study examining patients 8 months after COVID-19 led the authors to conclude that "B cell memory to SARS-CoV-2 was robust and is likely long-lasting." Reason #2 tells us that memory B cells can be active for a very long time indeed.
REASON #2: Memory B Cells Can Produce Neutralizing Antibodies If They See Infection Again Decades Later
Demonstrated production of memory B cells after vaccination or natural infection with COVID-19 is so important because memory B cells, once generated, can be activated to produce high levels of neutralizing antibodies against the pathogen even if encountered many years after the initial exposure. In one amazing study (published in 2008), researchers isolated memory B cells against the 1918 flu strain from the blood of 32 individuals aged 91-101 years. These people had been born on or before 1915 and had survived that pandemic.
Their memory B cells, when exposed to the 1918 flu strain in a test tube, generated high levels of neutralizing antibodies against the virus -- antibodies that then protected mice from lethal infection with this deadly strain. The ability of memory B cells to produce complex antibody responses against an infection nine decades after exposure speaks to their durability.
REASON #3: Vaccines or Natural Infection Trigger Strong Memory T Cell Immunity
All of the trials of the major COVID-19 vaccine candidates measured strong T cell immunity following vaccination, most often assessed by measuring SARS-CoV-2 specific T cells in the phase I/II safety and immunogenicity studies. There are a number of studies that demonstrate the production of strong T cell immunity to COVID-19 after natural infection as well, even when the infection was mild or asymptomatic.
The same study that showed us robust memory B cell production 8 months after natural infection also demonstrated strong and sustained memory T cell production. In fact, the half-lives of the memory T cells in this cohort were long (~125-225 days for CD8+ and ~94-153 days for CD4+ T cells), comparable to the 123-day half-life observed for memory CD8+ T cells after yellow fever immunization (a vaccine usually given once over a lifetime).
A recent study of individuals recovered from COVID-19 show that the initial T cells generated by natural infection mature and differentiate over time into memory T cells that will be "put in the bank" for sustained periods.
REASON #4: T Cell Immunity Following Vaccinations for Other Infections Is Long-Lasting
Last year, we were fortunate to be able to measure how T cell immunity is generated by COVID-19 vaccines, which was not possible in earlier eras when vaccine trials were done for other infections (such as measles, mumps, rubella, pertussis, diphtheria). Antibodies are just the "tip of the iceberg" when assessing the response to vaccination, but were the only arm of the immune response that could be measured following vaccination in the past.
Measuring pathogen-specific T cell responses takes sophisticated technology. However, T cell responses, when assessed years after vaccination for other pathogens, has been shown to be long-lasting. For example, in one study of 56 volunteers who had undergone measles vaccination when they were much younger, strong CD8 and CD4 cell responses to vaccination could be detected up to 34 years later.
REASON #5: T Cell Immunity to Related Coronaviruses That Caused Severe Disease is Long-Lasting
SARS-CoV-2 is a coronavirus that causes severe disease, unlike coronaviruses that cause the common cold. Two other coronaviruses in the recent past caused severe disease, specifically Severely Acute Respiratory Distress Syndrome (SARS) in late 2002-2003 and Middle East Respiratory Syndrome (MERS) in 2011.
A study performed in 2020 demonstrated that the blood of 23 recovered SARS patients possess long-lasting memory T cells that were still reactive to SARS 17 years after the outbreak in 2003. Many scientists expect that T cell immunity to SARS-CoV-2 will be equally durable to that of its cousin.
REASON #6: T Cell Responses from Vaccination and Natural Infection With the Ancestral Strain of COVID-19 Are Robust Against Variants
Even though antibody responses from vaccination may be slightly lower against various COVID-19 variants of concern that have emerged in recent months, T cell immunity after vaccination has been shown to be unperturbed by mutations in the spike protein (in the variants). For instance, T cell responses after mRNA vaccines maintained strong activity against different variants (including P.1 Brazil variant, B.1.1.7 UK variant, B.1.351 South Africa variant and the CA.20.C California variant) in a recent study.
Another study showed that the vaccines generated robust T cell immunity that was unfazed by different variants, including B.1.351 and B.1.1.7. The CD4 and CD8 responses generated after natural infection are equally robust, showing activity against multiple "epitopes" (little segments) of the spike protein of the virus. For instance, CD8 cells responds to 52 epitopes and CD4 cells respond to 57 epitopes across the spike protein, so that a few mutations in the variants cannot knock out such a robust and in-breadth T cell response. Indeed, a recent paper showed that mRNA vaccines were 97.4 percent effective against severe COVID-19 disease in Qatar, even when the majority of circulating virus there was from variants of concern (B.1.351 and B.1.1.7).
REASON #7: Coronaviruses Don't Mutate Quickly Like Influenza, Which Requires Annual Booster Shots
Coronaviruses are RNA viruses, like influenza and HIV (which is actually a retrovirus), but do not mutate as quickly as either one. The reason that coronaviruses don't mutate very rapidly is that their replicating mechanism (polymerase) has a strong proofreading mechanism: If the virus mutates, it usually goes back and self-corrects. Mutations can arise with high rates of replication when transmission is very frequent -- as has been seen in recent months with the emergence of SARS-CoV-2 variants during surges. However, the COVID-19 virus will not be mutating like this when we tamp down transmission with mass vaccination.
In conclusion, I and many of my infectious disease colleagues expect the immunity from natural infection or vaccination to COVID-19 to be durable. Let's put discussion of boosters aside and work hard on global vaccine equity and distribution since the pandemic is not over until it is over for us all.

