Genetically Sequencing Healthy Babies Yielded Surprising Results

A newborn and mother in the hospital - first touch. (© martin81/Shutterstock)
Today in Melrose, Massachusetts, Cora Stetson is the picture of good health, a bubbly precocious 2-year-old. But Cora has two separate mutations in the gene that produces a critical enzyme called biotinidase and her body produces only 40 percent of the normal levels of that enzyme.
In the last few years, the dream of predicting and preventing diseases through genomics, starting in childhood, is finally within reach.
That's enough to pass conventional newborn (heelstick) screening, but may not be enough for normal brain development, putting baby Cora at risk for seizures and cognitive impairment. But thanks to an experimental study in which Cora's DNA was sequenced after birth, this condition was discovered and she is being treated with a safe and inexpensive vitamin supplement.
Stories like these are beginning to emerge from the BabySeq Project, the first clinical trial in the world to systematically sequence healthy newborn infants. This trial was led by my research group with funding from the National Institutes of Health. While still controversial, it is pointing the way to a future in which adults, or even newborns, can receive comprehensive genetic analysis in order to determine their risk of future disease and enable opportunities to prevent them.
Some believe that medicine is still not ready for genomic population screening, but others feel it is long overdue. After all, the sequencing of the Human Genome Project was completed in 2003, and with this milestone, it became feasible to sequence and interpret the genome of any human being. The costs have come down dramatically since then; an entire human genome can now be sequenced for about $800, although the costs of bioinformatic and medical interpretation can add another $200 to $2000 more, depending upon the number of genes interrogated and the sophistication of the interpretive effort.

Two-year-old Cora Stetson, whose DNA sequencing after birth identified a potentially dangerous genetic mutation in time for her to receive preventive treatment.
(Photo courtesy of Robert Green)
The ability to sequence the human genome yielded extraordinary benefits in scientific discovery, disease diagnosis, and targeted cancer treatment. But the ability of genomes to detect health risks in advance, to actually predict the medical future of an individual, has been mired in controversy and slow to manifest. In particular, the oft-cited vision that healthy infants could be genetically tested at birth in order to predict and prevent the diseases they would encounter, has proven to be far tougher to implement than anyone anticipated.
But in the last few years, the dream of predicting and preventing diseases through genomics, starting in childhood, is finally within reach. Why did it take so long? And what remains to be done?
Great Expectations
Part of the problem was the unrealistic expectations that had been building for years in advance of the genomic science itself. For example, the 1997 film Gattaca portrayed a near future in which the lifetime risk of disease was readily predicted the moment an infant is born. In the fanfare that accompanied the completion of the Human Genome Project, the notion of predicting and preventing future disease in an individual became a powerful meme that was used to inspire investment and public support for genomic research long before the tools were in place to make it happen.
Another part of the problem was the success of state-mandated newborn screening programs that began in the 1960's with biochemical tests of the "heel-stick" for babies with metabolic disorders. These programs have worked beautifully, costing only a few dollars per baby and saving thousands of infants from death and severe cognitive impairment. It seemed only logical that a new technology like genome sequencing would add power and promise to such programs. But instead of embracing the notion of newborn sequencing, newborn screening laboratories have thus far rejected the entire idea as too expensive, too ambiguous, and too threatening to the comfortable constituency that they had built within the public health framework.
"What can you find when you look as deeply as possible into the medical genomes of healthy individuals?"
Creating the Evidence Base for Preventive Genomics
Despite a number of obstacles, there are researchers who are exploring how to achieve the original vision of genomic testing as a tool for disease prediction and prevention. For example, in our NIH-funded MedSeq Project, we were the first to ask the question: "What can you find when you look as deeply as possible into the medical genomes of healthy individuals?"
Most people do not understand that genetic information comes in four separate categories: 1) dominant mutations putting the individual at risk for rare conditions like familial forms of heart disease or cancer, (2) recessive mutations putting the individual's children at risk for rare conditions like cystic fibrosis or PKU, (3) variants across the genome that can be tallied to construct polygenic risk scores for common conditions like heart disease or type 2 diabetes, and (4) variants that can influence drug metabolism or predict drug side effects such as the muscle pain that occasionally occurs with statin use.
The technological and analytical challenges of our study were formidable, because we decided to systematically interrogate over 5000 disease-associated genes and report results in all four categories of genetic information directly to the primary care physicians for each of our volunteers. We enrolled 200 adults and found that everyone who was sequenced had medically relevant polygenic and pharmacogenomic results, over 90 percent carried recessive mutations that could have been important to reproduction, and an extraordinary 14.5 percent carried dominant mutations for rare genetic conditions.
A few years later we launched the BabySeq Project. In this study, we restricted the number of genes to include only those with child/adolescent onset that could benefit medically from early warning, and even so, we found 9.4 percent carried dominant mutations for rare conditions.
At first, our interpretation around the high proportion of apparently healthy individuals with dominant mutations for rare genetic conditions was simple – that these conditions had lower "penetrance" than anticipated; in other words, only a small proportion of those who carried the dominant mutation would get the disease. If this interpretation were to hold, then genetic risk information might be far less useful than we had hoped.
Suddenly the information available in the genome of even an apparently healthy individual is looking more robust, and the prospect of preventive genomics is looking feasible.
But then we circled back with each adult or infant in order to examine and test them for any possible features of the rare disease in question. When we did this, we were surprised to see that in over a quarter of those carrying such mutations, there were already subtle signs of the disease in question that had not even been suspected! Now our interpretation was different. We now believe that genetic risk may be responsible for subclinical disease in a much higher proportion of people than has ever been suspected!
Meanwhile, colleagues of ours have been demonstrating that detailed analysis of polygenic risk scores can identify individuals at high risk for common conditions like heart disease. So adding up the medically relevant results in any given genome, we start to see that you can learn your risks for a rare monogenic condition, a common polygenic condition, a bad effect from a drug you might take in the future, or for having a child with a devastating recessive condition. Suddenly the information available in the genome of even an apparently healthy individual is looking more robust, and the prospect of preventive genomics is looking feasible.
Preventive Genomics Arrives in Clinical Medicine
There is still considerable evidence to gather before we can recommend genomic screening for the entire population. For example, it is important to make sure that families who learn about such risks do not suffer harms or waste resources from excessive medical attention. And many doctors don't yet have guidance on how to use such information with their patients. But our research is convincing many people that preventive genomics is coming and that it will save lives.
In fact, we recently launched a Preventive Genomics Clinic at Brigham and Women's Hospital where information-seeking adults can obtain predictive genomic testing with the highest quality interpretation and medical context, and be coached over time in light of their disease risks toward a healthier outcome. Insurance doesn't yet cover such testing, so patients must pay out of pocket for now, but they can choose from a menu of genetic screening tests, all of which are more comprehensive than consumer-facing products. Genetic counseling is available but optional. So far, this service is for adults only, but sequencing for children will surely follow soon.
As the costs of sequencing and other Omics technologies continue to decline, we will see both responsible and irresponsible marketing of genetic testing, and we will need to guard against unscientific claims. But at the same time, we must be far more imaginative and fast moving in mainstream medicine than we have been to date in order to claim the emerging benefits of preventive genomics where it is now clear that suffering can be averted, and lives can be saved. The future has arrived if we are bold enough to grasp it.
Funding and Disclosures:
Dr. Green's research is supported by the National Institutes of Health, the Department of Defense and through donations to The Franca Sozzani Fund for Preventive Genomics. Dr. Green receives compensation for advising the following companies: AIA, Applied Therapeutics, Helix, Ohana, OptraHealth, Prudential, Verily and Veritas; and is co-founder and advisor to Genome Medical, Inc, a technology and services company providing genetics expertise to patients, providers, employers and care systems.
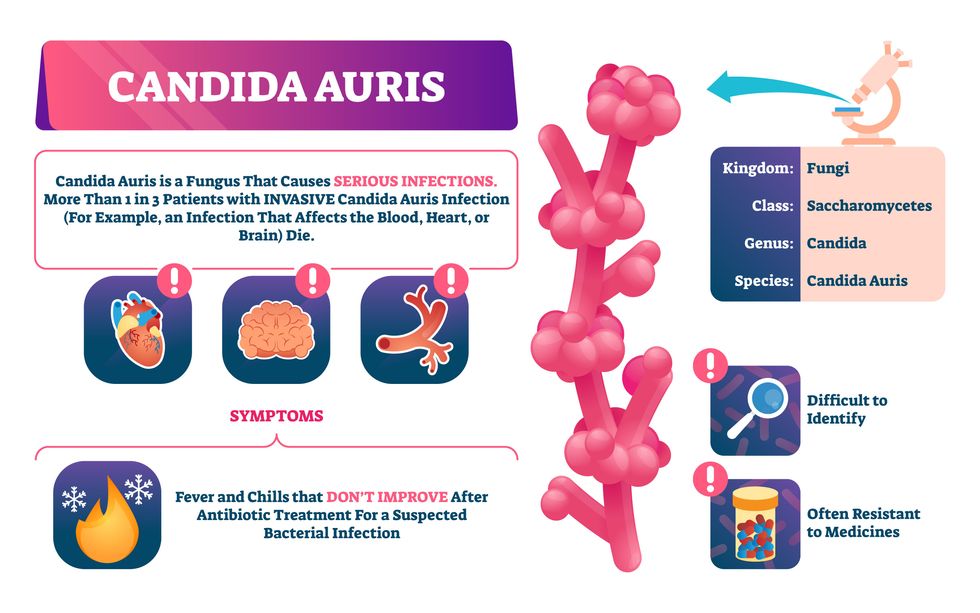
Doctors worry that fungal pathogens may cause the next pandemic.
Bacterial antibiotic resistance has been a concern in the medical field for several years. Now a new, similar threat is arising: drug-resistant fungal infections. The Centers for Disease Control and Prevention considers antifungal and antimicrobial resistance to be among the world’s greatest public health challenges.
One particular type of fungal infection caused by Candida auris is escalating rapidly throughout the world. And to make matters worse, C. auris is becoming increasingly resistant to current antifungal medications, which means that if you develop a C. auris infection, the drugs your doctor prescribes may not work. “We’re effectively out of medicines,” says Thomas Walsh, founding director of the Center for Innovative Therapeutics and Diagnostics, a translational research center dedicated to solving the antimicrobial resistance problem. Walsh spoke about the challenges at a Demy-Colton Virtual Salon, one in a series of interactive discussions among life science thought leaders.
Although C. auris typically doesn’t sicken healthy people, it afflicts immunocompromised hospital patients and may cause severe infections that can lead to sepsis, a life-threatening condition in which the overwhelmed immune system begins to attack the body’s own organs. Between 30 and 60 percent of patients who contract a C. auris infection die from it, according to the CDC. People who are undergoing stem cell transplants, have catheters or have taken antifungal or antibiotic medicines are at highest risk. “We’re coming to a perfect storm of increasing resistance rates, increasing numbers of immunosuppressed patients worldwide and a bug that is adapting to higher temperatures as the climate changes,” says Prabhavathi Fernandes, chair of the National BioDefense Science Board.
Most Candida species aren’t well-adapted to our body temperatures so they aren’t a threat. C. auris, however, thrives at human body temperatures.
Although medical professionals aren’t concerned at this point about C. auris evolving to affect healthy people, they worry that its presence in hospitals can turn routine surgeries into life-threatening calamities. “It’s coming,” says Fernandes. “It’s just a matter of time.”
An emerging global threat
“Fungi are found in the environment,” explains Fernandes, so Candida spores can easily wind up on people’s skin. In hospitals, they can be transferred from contact with healthcare workers or contaminated surfaces. Most Candida species aren’t well-adapted to our body temperatures so they aren’t a threat. C. auris, however, thrives at human body temperatures. It can enter the body during medical treatments that break the skin—and cause an infection. Overall, fungal infections cost some $48 billion in the U.S. each year. And infection rates are increasing because, in an ironic twist, advanced medical therapies are enabling severely ill patients to live longer and, therefore, be exposed to this pathogen.
The first-ever case of a C. auris infection was reported in Japan in 2009, although an analysis of Candida samples dated the earliest strain to a 1996 sample from South Korea. Since then, five separate varieties – called clades, which are similar to strains among bacteria – developed independently in different geographies: South Asia, East Asia, South Africa, South America and, recently, Iran. So far, C. auris infections have been reported in 35 countries.
In the U.S., the first infection was reported in 2016, and the CDC started tracking it nationally two years later. During that time, 5,654 cases have been reported to the CDC, which only tracks U.S. data.
What’s more notable than the number of cases is their rate of increase. In 2016, new cases increased by 175 percent and, on average, they have approximately doubled every year. From 2016 through 2022, the number of infections jumped from 63 to 2,377, a roughly 37-fold increase.
“This reminds me of what we saw with epidemics from 2013 through 2020… with Ebola, Zika and the COVID-19 pandemic,” says Robin Robinson, CEO of Spriovas and founding director of the Biomedical Advanced Research and Development Authority (BARDA), which is part of the U.S. Department of Health and Human Services. These epidemics started with a hockey stick trajectory, Robinson says—a gradual growth leading to a sharp spike, just like the shape of a hockey stick.
Another challenge is that right now medics don’t have rapid diagnostic tests for fungal infections. Currently, patients are often misdiagnosed because C. auris resembles several other easily treated fungi. Or they are diagnosed long after the infection begins and is harder to treat.
The problem is that existing diagnostics tests can only identify C. auris once it reaches the bloodstream. Yet, because this pathogen infects bodily tissues first, it should be possible to catch it much earlier before it becomes life-threatening. “We have to diagnose it before it reaches the bloodstream,” Walsh says.
The most alarming fact is that some Candida infections no longer respond to standard therapeutics.
“We need to focus on rapid diagnostic tests that do not rely on a positive blood culture,” says John Sperzel, president and CEO of T2 Biosystems, a company specializing in diagnostics solutions. Blood cultures typically take two to three days for the concentration of Candida to become large enough to detect. The company’s novel test detects about 90 percent of Candida species within three to five hours—thanks to its ability to spot minute quantities of the pathogen in blood samples instead of waiting for them to incubate and proliferate.
Unlike other Candida species C. auris thrives at human body temperatures
Adobe Stock
Tackling the resistance challenge
The most alarming fact is that some Candida infections no longer respond to standard therapeutics. The number of cases that stopped responding to echinocandin, the first-line therapy for most Candida infections, tripled in 2020, according to a study by the CDC.
Now, each of the first four clades shows varying levels of resistance to all three commonly prescribed classes of antifungal medications, such as azoles, echinocandins, and polyenes. For example, 97 percent of infections from C. auris Clade I are resistant to fluconazole, 54 percent to voriconazole and 30 percent of amphotericin. Nearly half are resistant to multiple antifungal drugs. Even with Clade II fungi, which has the least resistance of all the clades, 11 to 14 percent have become resistant to fluconazole.
Anti-fungal therapies typically target specific chemical compounds present on fungi’s cell membranes, but not on human cells—otherwise the medicine would cause damage to our own tissues. Fluconazole and other azole antifungals target a compound called ergosterol, preventing the fungal cells from replicating. Over the years, however, C. auris evolved to resist it, so existing fungal medications don’t work as well anymore.
A newer class of drugs called echinocandins targets a different part of the fungal cell. “The echinocandins – like caspofungin – inhibit (a part of the fungi) involved in making glucan, which is an essential component of the fungal cell wall and is not found in human cells,” Fernandes says. New antifungal treatments are needed, she adds, but there are only a few magic bullets that will hit just the fungus and not the human cells.
Research to fight infections also has been challenged by a lack of government support. That is changing now that BARDA is requesting proposals to develop novel antifungals. “The scope includes C. auris, as well as antifungals following a radiological/nuclear emergency, says BARDA spokesperson Elleen Kane.
The remaining challenge is the number of patients available to participate in clinical trials. Large numbers are needed, but the available patients are quite sick and often die before trials can be completed. Consequently, few biopharmaceutical companies are developing new treatments for C. auris.
ClinicalTrials.gov reports only two drugs in development for invasive C. auris infections—those than can spread throughout the body rather than localize in one particular area, like throat or vaginal infections: ibrexafungerp by Scynexis, Inc., fosmanogepix, by Pfizer.
Scynexis’ ibrexafungerp appears active against C. auris and other emerging, drug-resistant pathogens. The FDA recently approved it as a therapy for vaginal yeast infections and it is undergoing Phase III clinical trials against invasive candidiasis in an attempt to keep the infection from spreading.
“Ibreafungerp is structurally different from other echinocandins,” Fernandes says, because it targets a different part of the fungus. “We’re lucky it has activity against C. auris.”
Pfizer’s fosmanogepix is in Phase II clinical trials for patients with invasive fungal infections caused by multiple Candida species. Results are showing significantly better survival rates for people taking fosmanogepix.
Although C. auris does pose a serious threat to healthcare worldwide, scientists try to stay optimistic—because they recognized the problem early enough, they might have solutions in place before the perfect storm hits. “There is a bit of hope,” says Robinson. “BARDA has finally been able to fund the development of new antifungal agents and, hopefully, this year we can get several new classes of antifungals into development.”
New elevators could lift up our access to space
A space elevator would be cheaper and cleaner than using rockets
Story by Big Think
When people first started exploring space in the 1960s, it cost upwards of $80,000 (adjusted for inflation) to put a single pound of payload into low-Earth orbit.
A major reason for this high cost was the need to build a new, expensive rocket for every launch. That really started to change when SpaceX began making cheap, reusable rockets, and today, the company is ferrying customer payloads to LEO at a price of just $1,300 per pound.
This is making space accessible to scientists, startups, and tourists who never could have afforded it previously, but the cheapest way to reach orbit might not be a rocket at all — it could be an elevator.
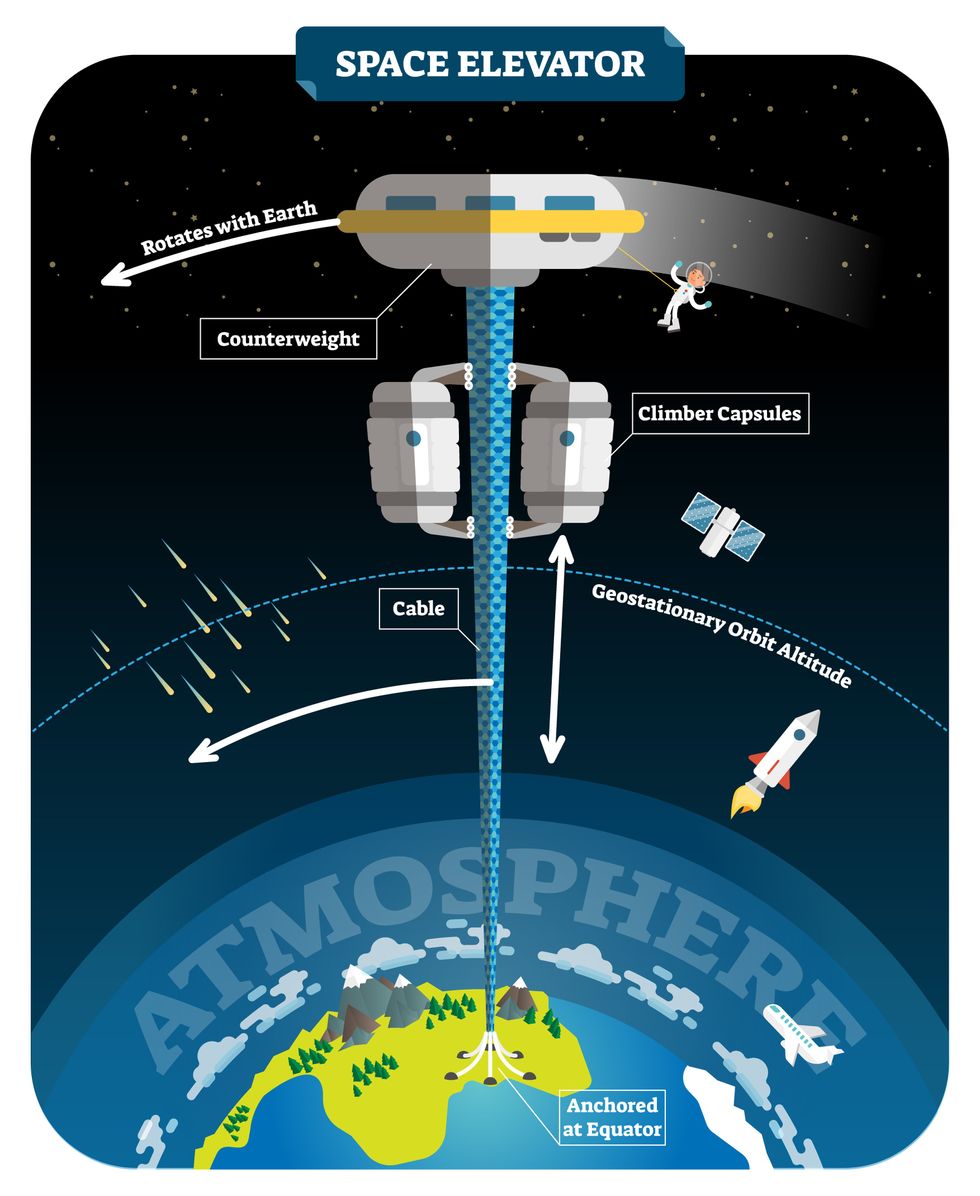
The space elevator
The seeds for a space elevator were first planted by Russian scientist Konstantin Tsiolkovsky in 1895, who, after visiting the 1,000-foot (305 m) Eiffel Tower, published a paper theorizing about the construction of a structure 22,000 miles (35,400 km) high.
This would provide access to geostationary orbit, an altitude where objects appear to remain fixed above Earth’s surface, but Tsiolkovsky conceded that no material could support the weight of such a tower.
We could then send electrically powered “climber” vehicles up and down the tether to deliver payloads to any Earth orbit.
In 1959, soon after Sputnik, Russian engineer Yuri N. Artsutanov proposed a way around this issue: instead of building a space elevator from the ground up, start at the top. More specifically, he suggested placing a satellite in geostationary orbit and dropping a tether from it down to Earth’s equator. As the tether descended, the satellite would ascend. Once attached to Earth’s surface, the tether would be kept taut, thanks to a combination of gravitational and centrifugal forces.
We could then send electrically powered “climber” vehicles up and down the tether to deliver payloads to any Earth orbit. According to physicist Bradley Edwards, who researched the concept for NASA about 20 years ago, it’d cost $10 billion and take 15 years to build a space elevator, but once operational, the cost of sending a payload to any Earth orbit could be as low as $100 per pound.
“Once you reduce the cost to almost a Fed-Ex kind of level, it opens the doors to lots of people, lots of countries, and lots of companies to get involved in space,” Edwards told Space.com in 2005.
In addition to the economic advantages, a space elevator would also be cleaner than using rockets — there’d be no burning of fuel, no harmful greenhouse emissions — and the new transport system wouldn’t contribute to the problem of space junk to the same degree that expendable rockets do.
So, why don’t we have one yet?
Tether troubles
Edwards wrote in his report for NASA that all of the technology needed to build a space elevator already existed except the material needed to build the tether, which needs to be light but also strong enough to withstand all the huge forces acting upon it.
The good news, according to the report, was that the perfect material — ultra-strong, ultra-tiny “nanotubes” of carbon — would be available in just two years.
“[S]teel is not strong enough, neither is Kevlar, carbon fiber, spider silk, or any other material other than carbon nanotubes,” wrote Edwards. “Fortunately for us, carbon nanotube research is extremely hot right now, and it is progressing quickly to commercial production.”Unfortunately, he misjudged how hard it would be to synthesize carbon nanotubes — to date, no one has been able to grow one longer than 21 inches (53 cm).
Further research into the material revealed that it tends to fray under extreme stress, too, meaning even if we could manufacture carbon nanotubes at the lengths needed, they’d be at risk of snapping, not only destroying the space elevator, but threatening lives on Earth.
Looking ahead
Carbon nanotubes might have been the early frontrunner as the tether material for space elevators, but there are other options, including graphene, an essentially two-dimensional form of carbon that is already easier to scale up than nanotubes (though still not easy).
Contrary to Edwards’ report, Johns Hopkins University researchers Sean Sun and Dan Popescu say Kevlar fibers could work — we would just need to constantly repair the tether, the same way the human body constantly repairs its tendons.
“Using sensors and artificially intelligent software, it would be possible to model the whole tether mathematically so as to predict when, where, and how the fibers would break,” the researchers wrote in Aeon in 2018.
“When they did, speedy robotic climbers patrolling up and down the tether would replace them, adjusting the rate of maintenance and repair as needed — mimicking the sensitivity of biological processes,” they continued.Astronomers from the University of Cambridge and Columbia University also think Kevlar could work for a space elevator — if we built it from the moon, rather than Earth.
They call their concept the Spaceline, and the idea is that a tether attached to the moon’s surface could extend toward Earth’s geostationary orbit, held taut by the pull of our planet’s gravity. We could then use rockets to deliver payloads — and potentially people — to solar-powered climber robots positioned at the end of this 200,000+ mile long tether. The bots could then travel up the line to the moon’s surface.
This wouldn’t eliminate the need for rockets to get into Earth’s orbit, but it would be a cheaper way to get to the moon. The forces acting on a lunar space elevator wouldn’t be as strong as one extending from Earth’s surface, either, according to the researchers, opening up more options for tether materials.
“[T]he necessary strength of the material is much lower than an Earth-based elevator — and thus it could be built from fibers that are already mass-produced … and relatively affordable,” they wrote in a paper shared on the preprint server arXiv.
After riding up the Earth-based space elevator, a capsule would fly to a space station attached to the tether of the moon-based one.
Electrically powered climber capsules could go up down the tether to deliver payloads to any Earth orbit.
Adobe Stock
Some Chinese researchers, meanwhile, aren’t giving up on the idea of using carbon nanotubes for a space elevator — in 2018, a team from Tsinghua University revealed that they’d developed nanotubes that they say are strong enough for a tether.
The researchers are still working on the issue of scaling up production, but in 2021, state-owned news outlet Xinhua released a video depicting an in-development concept, called “Sky Ladder,” that would consist of space elevators above Earth and the moon.
After riding up the Earth-based space elevator, a capsule would fly to a space station attached to the tether of the moon-based one. If the project could be pulled off — a huge if — China predicts Sky Ladder could cut the cost of sending people and goods to the moon by 96 percent.
The bottom line
In the 120 years since Tsiolkovsky looked at the Eiffel Tower and thought way bigger, tremendous progress has been made developing materials with the properties needed for a space elevator. At this point, it seems likely we could one day have a material that can be manufactured at the scale needed for a tether — but by the time that happens, the need for a space elevator may have evaporated.
Several aerospace companies are making progress with their own reusable rockets, and as those join the market with SpaceX, competition could cause launch prices to fall further.
California startup SpinLaunch, meanwhile, is developing a massive centrifuge to fling payloads into space, where much smaller rockets can propel them into orbit. If the company succeeds (another one of those big ifs), it says the system would slash the amount of fuel needed to reach orbit by 70 percent.
Even if SpinLaunch doesn’t get off the ground, several groups are developing environmentally friendly rocket fuels that produce far fewer (or no) harmful emissions. More work is needed to efficiently scale up their production, but overcoming that hurdle will likely be far easier than building a 22,000-mile (35,400-km) elevator to space.
This article originally appeared on Big Think, home of the brightest minds and biggest ideas of all time.


