Genetically Sequencing Healthy Babies Yielded Surprising Results

A newborn and mother in the hospital - first touch. (© martin81/Shutterstock)
Today in Melrose, Massachusetts, Cora Stetson is the picture of good health, a bubbly precocious 2-year-old. But Cora has two separate mutations in the gene that produces a critical enzyme called biotinidase and her body produces only 40 percent of the normal levels of that enzyme.
In the last few years, the dream of predicting and preventing diseases through genomics, starting in childhood, is finally within reach.
That's enough to pass conventional newborn (heelstick) screening, but may not be enough for normal brain development, putting baby Cora at risk for seizures and cognitive impairment. But thanks to an experimental study in which Cora's DNA was sequenced after birth, this condition was discovered and she is being treated with a safe and inexpensive vitamin supplement.
Stories like these are beginning to emerge from the BabySeq Project, the first clinical trial in the world to systematically sequence healthy newborn infants. This trial was led by my research group with funding from the National Institutes of Health. While still controversial, it is pointing the way to a future in which adults, or even newborns, can receive comprehensive genetic analysis in order to determine their risk of future disease and enable opportunities to prevent them.
Some believe that medicine is still not ready for genomic population screening, but others feel it is long overdue. After all, the sequencing of the Human Genome Project was completed in 2003, and with this milestone, it became feasible to sequence and interpret the genome of any human being. The costs have come down dramatically since then; an entire human genome can now be sequenced for about $800, although the costs of bioinformatic and medical interpretation can add another $200 to $2000 more, depending upon the number of genes interrogated and the sophistication of the interpretive effort.

Two-year-old Cora Stetson, whose DNA sequencing after birth identified a potentially dangerous genetic mutation in time for her to receive preventive treatment.
(Photo courtesy of Robert Green)
The ability to sequence the human genome yielded extraordinary benefits in scientific discovery, disease diagnosis, and targeted cancer treatment. But the ability of genomes to detect health risks in advance, to actually predict the medical future of an individual, has been mired in controversy and slow to manifest. In particular, the oft-cited vision that healthy infants could be genetically tested at birth in order to predict and prevent the diseases they would encounter, has proven to be far tougher to implement than anyone anticipated.
But in the last few years, the dream of predicting and preventing diseases through genomics, starting in childhood, is finally within reach. Why did it take so long? And what remains to be done?
Great Expectations
Part of the problem was the unrealistic expectations that had been building for years in advance of the genomic science itself. For example, the 1997 film Gattaca portrayed a near future in which the lifetime risk of disease was readily predicted the moment an infant is born. In the fanfare that accompanied the completion of the Human Genome Project, the notion of predicting and preventing future disease in an individual became a powerful meme that was used to inspire investment and public support for genomic research long before the tools were in place to make it happen.
Another part of the problem was the success of state-mandated newborn screening programs that began in the 1960's with biochemical tests of the "heel-stick" for babies with metabolic disorders. These programs have worked beautifully, costing only a few dollars per baby and saving thousands of infants from death and severe cognitive impairment. It seemed only logical that a new technology like genome sequencing would add power and promise to such programs. But instead of embracing the notion of newborn sequencing, newborn screening laboratories have thus far rejected the entire idea as too expensive, too ambiguous, and too threatening to the comfortable constituency that they had built within the public health framework.
"What can you find when you look as deeply as possible into the medical genomes of healthy individuals?"
Creating the Evidence Base for Preventive Genomics
Despite a number of obstacles, there are researchers who are exploring how to achieve the original vision of genomic testing as a tool for disease prediction and prevention. For example, in our NIH-funded MedSeq Project, we were the first to ask the question: "What can you find when you look as deeply as possible into the medical genomes of healthy individuals?"
Most people do not understand that genetic information comes in four separate categories: 1) dominant mutations putting the individual at risk for rare conditions like familial forms of heart disease or cancer, (2) recessive mutations putting the individual's children at risk for rare conditions like cystic fibrosis or PKU, (3) variants across the genome that can be tallied to construct polygenic risk scores for common conditions like heart disease or type 2 diabetes, and (4) variants that can influence drug metabolism or predict drug side effects such as the muscle pain that occasionally occurs with statin use.
The technological and analytical challenges of our study were formidable, because we decided to systematically interrogate over 5000 disease-associated genes and report results in all four categories of genetic information directly to the primary care physicians for each of our volunteers. We enrolled 200 adults and found that everyone who was sequenced had medically relevant polygenic and pharmacogenomic results, over 90 percent carried recessive mutations that could have been important to reproduction, and an extraordinary 14.5 percent carried dominant mutations for rare genetic conditions.
A few years later we launched the BabySeq Project. In this study, we restricted the number of genes to include only those with child/adolescent onset that could benefit medically from early warning, and even so, we found 9.4 percent carried dominant mutations for rare conditions.
At first, our interpretation around the high proportion of apparently healthy individuals with dominant mutations for rare genetic conditions was simple – that these conditions had lower "penetrance" than anticipated; in other words, only a small proportion of those who carried the dominant mutation would get the disease. If this interpretation were to hold, then genetic risk information might be far less useful than we had hoped.
Suddenly the information available in the genome of even an apparently healthy individual is looking more robust, and the prospect of preventive genomics is looking feasible.
But then we circled back with each adult or infant in order to examine and test them for any possible features of the rare disease in question. When we did this, we were surprised to see that in over a quarter of those carrying such mutations, there were already subtle signs of the disease in question that had not even been suspected! Now our interpretation was different. We now believe that genetic risk may be responsible for subclinical disease in a much higher proportion of people than has ever been suspected!
Meanwhile, colleagues of ours have been demonstrating that detailed analysis of polygenic risk scores can identify individuals at high risk for common conditions like heart disease. So adding up the medically relevant results in any given genome, we start to see that you can learn your risks for a rare monogenic condition, a common polygenic condition, a bad effect from a drug you might take in the future, or for having a child with a devastating recessive condition. Suddenly the information available in the genome of even an apparently healthy individual is looking more robust, and the prospect of preventive genomics is looking feasible.
Preventive Genomics Arrives in Clinical Medicine
There is still considerable evidence to gather before we can recommend genomic screening for the entire population. For example, it is important to make sure that families who learn about such risks do not suffer harms or waste resources from excessive medical attention. And many doctors don't yet have guidance on how to use such information with their patients. But our research is convincing many people that preventive genomics is coming and that it will save lives.
In fact, we recently launched a Preventive Genomics Clinic at Brigham and Women's Hospital where information-seeking adults can obtain predictive genomic testing with the highest quality interpretation and medical context, and be coached over time in light of their disease risks toward a healthier outcome. Insurance doesn't yet cover such testing, so patients must pay out of pocket for now, but they can choose from a menu of genetic screening tests, all of which are more comprehensive than consumer-facing products. Genetic counseling is available but optional. So far, this service is for adults only, but sequencing for children will surely follow soon.
As the costs of sequencing and other Omics technologies continue to decline, we will see both responsible and irresponsible marketing of genetic testing, and we will need to guard against unscientific claims. But at the same time, we must be far more imaginative and fast moving in mainstream medicine than we have been to date in order to claim the emerging benefits of preventive genomics where it is now clear that suffering can be averted, and lives can be saved. The future has arrived if we are bold enough to grasp it.
Funding and Disclosures:
Dr. Green's research is supported by the National Institutes of Health, the Department of Defense and through donations to The Franca Sozzani Fund for Preventive Genomics. Dr. Green receives compensation for advising the following companies: AIA, Applied Therapeutics, Helix, Ohana, OptraHealth, Prudential, Verily and Veritas; and is co-founder and advisor to Genome Medical, Inc, a technology and services company providing genetics expertise to patients, providers, employers and care systems.
Why you should (virtually) care
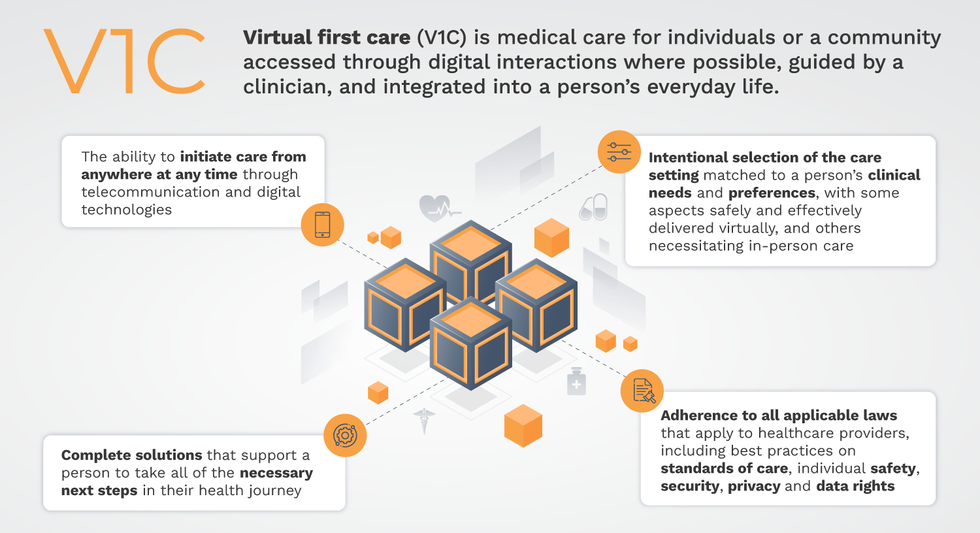
Virtual-first care, or V1C, could increase the quality of healthcare and make it more patient-centric by letting patients combine in-person visits with virtual options such as video for seeing their care providers.
As the pandemic turns endemic, healthcare providers have been eagerly urging patients to return to their offices to enjoy the benefits of in-person care.
But wait.
The last two years have forced all sorts of organizations to be nimble, adaptable and creative in how they work, and this includes healthcare providers’ efforts to maintain continuity of care under the most challenging of conditions. So before we go back to “business as usual,” don’t we owe it to those providers and ourselves to admit that business as usual did not work for most of the people the industry exists to help? If we’re going to embrace yet another period of change – periods that don’t happen often in our complex industry – shouldn’t we first stop and ask ourselves what we’re trying to achieve?
Certainly, COVID has shown that telehealth can be an invaluable tool, particularly for patients in rural and underserved communities that lack access to specialty care. It’s also become clear that many – though not all – healthcare encounters can be effectively conducted from afar. That said, the telehealth tactics that filled the gap during the pandemic were largely stitched together substitutes for existing visit-based workflows: with offices closed, patients scheduled video visits for help managing the side effects of their blood pressure medications or to see their endocrinologist for a quarterly check-in. Anyone whose children slogged through the last year or two of remote learning can tell you that simply virtualizing existing processes doesn’t necessarily improve the experience or the outcomes!
But what if our approach to post-pandemic healthcare came from a patient-driven perspective? We have a fleeting opportunity to advance a care model centered on convenient and equitable access that first prioritizes good outcomes, then selects approaches to care – and locations – tailored to each patient. Using the example of education, imagine how effective it would be if each student, regardless of their school district and aptitude, received such individualized attention.
That’s the idea behind virtual-first care (V1C), a new care model centered on convenient, customized, high-quality care that integrates a full suite of services tailored directly to patients’ clinical needs and preferences. This package includes asynchronous communication such as texting; video and other live virtual modes; and in-person options.
V1C goes beyond what you might think of as standard “telehealth” by using evidence-based protocols and tools that include traditional and digital therapeutics and testing, personalized care plans, dynamic patient monitoring, and team-based approaches to care. This could include spit kits mailed for laboratory tests and complementing clinical care with health coaching. V1C also replaces some in-person exams with ongoing monitoring, using sensors for more ‘whole person’ care.
Amidst all this momentum, we have the opportunity to rethink the goals of healthcare innovation, but that means bringing together key stakeholders to demonstrate that sustainable V1C can redefine healthcare.
Established V1C healthcare providers such as Omada, Headspace, and Heartbeat Health, as well as emerging market entrants like Oshi, Visana, and Wellinks, work with a variety of patients who have complicated long-term conditions such as diabetes, heart failure, gastrointestinal illness, endometriosis, and COPD. V1C is comprehensive in ways that are lacking in digital health and its other predecessors: it has the potential to integrate multiple data streams, incorporate more frequent touches and check-ins over time, and manage a much wider range of chronic health conditions, improving lives and reducing disease burden now and in the future.
Recognizing the pandemic-driven interest in virtual care, significant energy and resources are already flowing fast toward V1C. Some of the world’s largest innovators jumped into V1C early on: Verily, Alphabet’s Life Sciences Company, launched Onduo in 2016 to disrupt the diabetes healthcare market, and is now well positioned to scale its solutions. Major insurers like Aetna and United now offer virtual-first plans to members, responding as organizations expand virtual options for employees. Amidst all this momentum, we have the opportunity to rethink the goals of healthcare innovation, but that means bringing together key stakeholders to demonstrate that sustainable V1C can redefine healthcare.
That was the immediate impetus for IMPACT, a consortium of V1C companies, investors, payers and patients founded last year to ensure access to high-quality, evidence-based V1C. Developed by our team at the Digital Medicine Society (DiMe) in collaboration with the American Telemedicine Association (ATA), IMPACT has begun to explore key issues that include giving patients more integrated experiences when accessing both virtual and brick-and-mortar care.
Digital Medicine Society
V1C is not, nor should it be, virtual-only care. In this new era of hybrid healthcare, success will be defined by how well providers help patients navigate the transitions. How do we smoothly hand a patient off from an onsite primary care physician to, say, a virtual cardiologist? How do we get information from a brick-and-mortar to a digital portal? How do you manage dataflow while still staying HIPAA compliant? There are many complex regulatory implications for these new models, as well as an evolving landscape in terms of privacy, security and interoperability. It will be no small task for groups like IMPACT to determine the best path forward.
None of these factors matter unless the industry can recruit and retain clinicians. Our field is facing an unprecedented workforce crisis. Traditional healthcare is making clinicians miserable, and COVID has only accelerated the trend of overworked, disenchanted healthcare workers leaving in droves. Clinicians want more interactions with patients, and fewer with computer screens – call it “More face time, less FaceTime.” No new model will succeed unless the industry can more efficiently deploy its talent – arguably its most scarce and precious resource. V1C can help with alleviating the increasing burden and frustration borne by individual physicians in today’s status quo.
In healthcare, new technological approaches inevitably provoke no shortage of skepticism. Past lessons from Silicon Valley-driven fixes have led to understandable cynicism. But V1C is a different breed of animal. By building healthcare around the patient, not the clinic, V1C can make healthcare work better for patients, payers and providers. We’re at a fork in the road: we can revert back to a broken sick-care system, or dig in and do the hard work of figuring out how this future-forward healthcare system gets financed, organized and executed. As a field, we must find the courage and summon the energy to embrace this moment, and make it a moment of change.
Podcast: The future of brain health with Percy Griffin
Percy Griffin, director of scientific engagement for the Alzheimer’s Association, joins Leaps.org to discuss the present and future of the fight against dementia.
Today's guest is Percy Griffin, director of scientific engagement for the Alzheimer’s Association, a nonprofit that’s focused on speeding up research, finding better ways to detect Alzheimer’s earlier and other approaches for reducing risk. Percy has a doctorate in molecular cell biology from Washington University, he’s led important research on Alzheimer’s, and you can find the link to his full bio in the show notes, below.
Our topic for this conversation is the present and future of the fight against dementia. Billions of dollars have been spent by the National Institutes of Health and biotechs to research new treatments for Alzheimer's and other forms of dementia, but so far there's been little to show for it. Last year, Aduhelm became the first drug to be approved by the FDA for Alzheimer’s in 20 years, but it's received a raft of bad publicity, with red flags about its effectiveness, side effects and cost.
Meanwhile, 6.5 million Americans have Alzheimer's, and this number could increase to 13 million in 2050. Listen to this conversation if you’re concerned about your own brain health, that of family members getting older, or if you’re just concerned about the future of this country with experts predicting the number people over 65 will increase dramatically in the very near future.
Listen to the Episode
Listen on Apple | Listen on Spotify | Listen on Stitcher | Listen on Amazon | Listen on Google
4:40 - We talk about the parts of Percy’s life that led to him to concentrate on working in this important area.
6:20 - He defines Alzheimer's and dementia, and discusses the key elements of communicating science.
10:20 - Percy explains why the Alzheimer’s Association has been supportive of Aduhelm, even as others have been critical.
17:58 - We talk about therapeutics under development, which ones to be excited about, and how they could be tailored to a person's own biology.
24:25 - Percy discusses funding and tradeoffs between investing more money into Alzheimer’s research compared to other intractable diseases like cancer, and new opportunities to accelerate progress, such as ARPA-H, President Biden’s proposed agency to speed up health breakthroughs.
27:24 - We talk about the social determinants of brain health. What are the pros/cons of continuing to spend massive sums of money to develop new drugs like Aduhelm versus refocusing on expanding policies to address social determinants - like better education, nutritious food and safe drinking water - that have enabled some groups more than others to enjoy improved cognition late in life.
34:18 - Percy describes his top lifestyle recommendations for protecting your mind.
37:33 - Is napping bad for the brain?
39:39 - Circadian rhythm and Alzheimer's.
42:34 - What tests can people take to check their brain health today, and which biomarkers are we making progress on?
47:25 - Percy highlights important programs run by the Alzheimer’s Association to support advances.
Show links:
** After this episode was recorded, the Centers for Medicare and Medicaid Services affirmed its decision from last June to limit coverage of Aduhelm. More here.
- Percy Griffin's bio: https://www.alz.org/manh/events/alztalks/upcoming-...
- The Alzheimer's Association's Part the Cloud program: https://alz.org/partthecloud/about-us.asp
- The paradox of dementia rates decreasing: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7455342/
- The argument for focusing more resources on improving institutions and social processes for brain health: https://www.statnews.com/2021/09/23/the-brain-heal...
- Recent research on napping: https://www.ocregister.com/2022/03/25/alzheimers-s...
- The Alzheimer's Association helpline: https://www.alz.org/help-support/resources/helpline
- ALZConnected, a free online community for people affected by dementia https://www.alzconnected.org/
- TrialMatch for people with dementia and healthy volunteers to find clinical trials for Alzheimer's and other dementia: https://www.alz.org/alzheimers-dementia/research_p...

