What will the $100 genome mean?

A company has slashed the cost of assessing a person's genome to just $100. With lower costs - and as other genetic tools mature and evolve - a wave of new therapies could be coming in the near future.
In May 2022, Californian biotech Ultima Genomics announced that its UG 100 platform was capable of sequencing an entire human genome for just $100, a landmark moment in the history of the field. The announcement was particularly remarkable because few had previously heard of the company, a relative unknown in an industry long dominated by global giant Illumina which controls about 80 percent of the world’s sequencing market.
Ultima’s secret was to completely revamp many technical aspects of the way Illumina have traditionally deciphered DNA. The process usually involves first splitting the double helix DNA structure into single strands, then breaking these strands into short fragments which are laid out on a glass surface called a flow cell. When this flow cell is loaded into the sequencing machine, color-coded tags are attached to each individual base letter. A laser scans the bases individually while a camera simultaneously records the color associated with them, a process which is repeated until every single fragment has been sequenced.
Instead, Ultima has found a series of shortcuts to slash the cost and boost efficiency. “Ultima Genomics has developed a fundamentally new sequencing architecture designed to scale beyond conventional approaches,” says Josh Lauer, Ultima’s chief commercial officer.
This ‘new architecture’ is a series of subtle but highly impactful tweaks to the sequencing process ranging from replacing the costly flow cell with a silicon wafer which is both cheaper and allows more DNA to be read at once, to utilizing machine learning to convert optical data into usable information.
To put $100 genome in perspective, back in 2012 the cost of sequencing a single genome was around $10,000, a price tag which dropped to $1,000 a few years later. Before Ultima’s announcement, the cost of sequencing an individual genome was around $600.
Several studies have found that nearly 12 percent of healthy people who have their genome sequenced, then discover they have a variant pointing to a heightened risk of developing a disease that can be monitored, treated or prevented.
While Ultima’s new machine is not widely available yet, Illumina’s response has been rapid. In September 2022, the company unveiled the NovaSeq X series, which it describes as its fastest most cost-efficient sequencing platform yet, capable of sequencing genomes at $200, with further price cuts likely to follow.
But what will the rapidly tumbling cost of sequencing actually mean for medicine? “Well to start with, obviously it’s going to mean more people getting their genome sequenced,” says Michael Snyder, professor of genetics at Stanford University. “It'll be a lot more accessible to people.”
At the moment sequencing is mainly limited to certain cancer patients where it is used to inform treatment options, and individuals with undiagnosed illnesses. In the past, initiatives such as SeqFirst have attempted further widen access to genome sequencing based on growing amounts of research illustrating the potential benefits of the technology in healthcare. Several studies have found that nearly 12 percent of healthy people who have their genome sequenced, then discover they have a variant pointing to a heightened risk of developing a disease that can be monitored, treated or prevented.
“While whole genome sequencing is not yet widely used in the U.S., it has started to come into pediatric critical care settings such as newborn intensive care units,” says Professor Michael Bamshad, who heads the genetic medicine division in the University of Washington’s pediatrics department. “It is also being used more often in outpatient clinical genetics services, particularly when conventional testing fails to identify explanatory variants.”
But the cost of sequencing itself is only one part of the price tag. The subsequent clinical interpretation and genetic counselling services often come to several thousand dollars, a cost which insurers are not always willing to pay.
As a result, while Bamshad and others hope that the arrival of the $100 genome will create new opportunities to use genetic testing in innovative ways, the most immediate benefits are likely to come in the realm of research.
Bigger Data
There are numerous ways in which cheaper sequencing is likely to advance scientific research, for example the ability to collect data on much larger patient groups. This will be a major boon to scientists working on complex heterogeneous diseases such as schizophrenia or depression where there are many genes involved which all exert subtle effects, as well as substantial variance across the patient population. Bigger studies could help scientists identify subgroups of patients where the disease appears to be driven by similar gene variants, who can then be more precisely targeted with specific drugs.
If insurers can figure out the economics, Snyder even foresees a future where at a certain age, all of us can qualify for annual sequencing of our blood cells to search for early signs of cancer or the potential onset of other diseases like type 2 diabetes.
David Curtis, a genetics professor at University College London, says that scientists studying these illnesses have previously been forced to rely on genome-wide association studies which are limited because they only identify common gene variants. “We might see a significant increase in the number of large association studies using sequence data,” he says. “It would be far preferable to use this because it provides information about rare, potentially functional variants.”
Cheaper sequencing will also aid researchers working on diseases which have traditionally been underfunded. Bamshad cites cystic fibrosis, a condition which affects around 40,000 children and adults in the U.S., as one particularly pertinent example.
“Funds for gene discovery for rare diseases are very limited,” he says. “We’re one of three sites that did whole genome sequencing on 5,500 people with cystic fibrosis, but our statistical power is limited. A $100 genome would make it much more feasible to sequence everyone in the U.S. with cystic fibrosis and make it more likely that we discover novel risk factors and pathways influencing clinical outcomes.”
For progressive diseases that are more common like cancer and type 2 diabetes, as well as neurodegenerative conditions like multiple sclerosis and ALS, geneticists will be able to go even further and afford to sequence individual tumor cells or neurons at different time points. This will enable them to analyze how individual DNA modifications like methylation, change as the disease develops.
In the case of cancer, this could help scientists understand how tumors evolve to evade treatments. Within in a clinical setting, the ability to sequence not just one, but many different cells across a patient’s tumor could point to the combination of treatments which offer the best chance of eradicating the entire cancer.
“What happens at the moment with a solid tumor is you treat with one drug, and maybe 80 percent of that tumor is susceptible to that drug,” says Neil Ward, vice president and general manager in the EMEA region for genomics company PacBio. “But the other 20 percent of the tumor has already got mutations that make it resistant, which is probably why a lot of modern therapies extend life for sadly only a matter of months rather than curing, because they treat a big percentage of the tumor, but not the whole thing. So going forwards, I think that we will see genomics play a huge role in cancer treatments, through using multiple modalities to treat someone's cancer.”
If insurers can figure out the economics, Snyder even foresees a future where at a certain age, all of us can qualify for annual sequencing of our blood cells to search for early signs of cancer or the potential onset of other diseases like type 2 diabetes.
“There are companies already working on looking for cancer signatures in methylated DNA,” he says. “If it was determined that you had early stage cancer, pre-symptomatically, that could then be validated with targeted MRI, followed by surgery or chemotherapy. It makes a big difference catching cancer early. If there were signs of type 2 diabetes, you could start taking steps to mitigate your glucose rise, and possibly prevent it or at least delay the onset.”
This would already revolutionize the way we seek to prevent a whole range of illnesses, but others feel that the $100 genome could also usher in even more powerful and controversial preventative medicine schemes.
Newborn screening
In the eyes of Kári Stefánsson, the Icelandic neurologist who been a visionary for so many advances in the field of human genetics over the last 25 years, the falling cost of sequencing means it will be feasible to sequence the genomes of every baby born.
“We have recently done an analysis of genomes in Iceland and the UK Biobank, and in 4 percent of people you find mutations that lead to serious disease, that can be prevented or dealt with,” says Stefansson, CEO of deCODE genetics, a subsidiary of the pharmaceutical company Amgen. “This could transform our healthcare systems.”
As well as identifying newborns with rare diseases, this kind of genomic information could be used to compute a person’s risk score for developing chronic illnesses later in life. If for example, they have a higher than average risk of colon or breast cancer, they could be pre-emptively scheduled for annual colonoscopies or mammograms as soon as they hit adulthood.
To a limited extent, this is already happening. In the UK, Genomics England has launched the Newborn Genomes Programme, which plans to undertake whole-genome sequencing of up to 200,000 newborn babies, with the aim of enabling the early identification of rare genetic diseases.
"I have not had my own genome sequenced and I would not have wanted my parents to have agreed to this," Curtis says. "I don’t see that sequencing children for the sake of some vague, ill-defined benefits could ever be justifiable.”
However, some scientists feel that it is tricky to justify sequencing the genomes of apparently healthy babies, given the data privacy issues involved. They point out that we still know too little about the links which can be drawn between genetic information at birth, and risk of chronic illness later in life.
“I think there are very difficult ethical issues involved in sequencing children if there are no clear and immediate clinical benefits,” says Curtis. “They cannot consent to this process. I have not had my own genome sequenced and I would not have wanted my parents to have agreed to this. I don’t see that sequencing children for the sake of some vague, ill-defined benefits could ever be justifiable.”
Curtis points out that there are many inherent risks about this data being available. It may fall into the hands of insurance companies, and it could even be used by governments for surveillance purposes.
“Genetic sequence data is very useful indeed for forensic purposes. Its full potential has yet to be realized but identifying rare variants could provide a quick and easy way to find relatives of a perpetrator,” he says. “If large numbers of people had been sequenced in a healthcare system then it could be difficult for a future government to resist the temptation to use this as a resource to investigate serious crimes.”
While sequencing becoming more widely available will present difficult ethical and moral challenges, it will offer many benefits for society as a whole. Cheaper sequencing will help boost the diversity of genomic datasets which have traditionally been skewed towards individuals of white, European descent, meaning that much of the actionable medical information which has come out of these studies is not relevant to people of other ethnicities.
Ward predicts that in the coming years, the growing amount of genetic information will ultimately change the outcomes for many with rare, previously incurable illnesses.
“If you're the parent of a child that has a susceptible or a suspected rare genetic disease, their genome will get sequenced, and while sadly that doesn’t always lead to treatments, it’s building up a knowledge base so companies can spring up and target that niche of a disease,” he says. “As a result there’s a whole tidal wave of new therapies that are going to come to market over the next five years, as the genetic tools we have, mature and evolve.”
This article was first published by Leaps.org in October 2022.
Nobel Prize goes to technology for mRNA vaccines
Katalin Karikó, pictured, and Drew Weissman won the Nobel Prize for advances in mRNA research that led to the first Covid vaccines.
When Drew Weissman received a call from Katalin Karikó in the early morning hours this past Monday, he assumed his longtime research partner was calling to share a nascent, nagging idea. Weissman, a professor of medicine at the Perelman School of Medicine at the University of Pennsylvania, and Karikó, a professor at Szeged University and an adjunct professor at UPenn, both struggle with sleep disturbances. Thus, middle-of-the-night discourses between the two, often over email, has been a staple of their friendship. But this time, Karikó had something more pressing and exciting to share: They had won the 2023 Nobel Prize in Physiology or Medicine.
The work for which they garnered the illustrious award and its accompanying $1,000,000 cash windfall was completed about two decades ago, wrought through long hours in the lab over many arduous years. But humanity collectively benefited from its life-saving outcome three years ago, when both Moderna and Pfizer/BioNTech’s mRNA vaccines against COVID were found to be safe and highly effective at preventing severe disease. Billions of doses have since been given out to protect humans from the upstart viral scourge.
“I thought of going somewhere else, or doing something else,” said Katalin Karikó. “I also thought maybe I’m not good enough, not smart enough. I tried to imagine: Everything is here, and I just have to do better experiments.”
Unlocking the power of mRNA
Weissman and Karikó unlocked mRNA vaccines for the world back in the early 2000s when they made a key breakthrough. Messenger RNA molecules are essentially instructions for cells’ ribosomes to make specific proteins, so in the 1980s and 1990s, researchers started wondering if sneaking mRNA into the body could trigger cells to manufacture antibodies, enzymes, or growth agents for protecting against infection, treating disease, or repairing tissues. But there was a big problem: injecting this synthetic mRNA triggered a dangerous, inflammatory immune response resulting in the mRNA’s destruction.
While most other researchers chose not to tackle this perplexing problem to instead pursue more lucrative and publishable exploits, Karikó stuck with it. The choice sent her academic career into depressing doldrums. Nobody would fund her work, publications dried up, and after six years as an assistant professor at the University of Pennsylvania, Karikó got demoted. She was going backward.
“I thought of going somewhere else, or doing something else,” Karikó told Stat in 2020. “I also thought maybe I’m not good enough, not smart enough. I tried to imagine: Everything is here, and I just have to do better experiments.”
A tale of tenacity
Collaborating with Drew Weissman, a new professor at the University of Pennsylvania, in the late 1990s helped provide Karikó with the tenacity to continue. Weissman nurtured a goal of developing a vaccine against HIV-1, and saw mRNA as a potential way to do it.
“For the 20 years that we’ve worked together before anybody knew what RNA is, or cared, it was the two of us literally side by side at a bench working together,” Weissman said in an interview with Adam Smith of the Nobel Foundation.
In 2005, the duo made their 2023 Nobel Prize-winning breakthrough, detailing it in a relatively small journal, Immunity. (Their paper was rejected by larger journals, including Science and Nature.) They figured out that chemically modifying the nucleoside bases that make up mRNA allowed the molecule to slip past the body’s immune defenses. Karikó and Weissman followed up that finding by creating mRNA that’s more efficiently translated within cells, greatly boosting protein production. In 2020, scientists at Moderna and BioNTech (where Karikó worked from 2013 to 2022) rushed to craft vaccines against COVID, putting their methods to life-saving use.
The future of vaccines
Buoyed by the resounding success of mRNA vaccines, scientists are now hurriedly researching ways to use mRNA medicine against other infectious diseases, cancer, and genetic disorders. The now ubiquitous efforts stand in stark contrast to Karikó and Weissman’s previously unheralded struggles years ago as they doggedly worked to realize a shared dream that so many others shied away from. Katalin Karikó and Drew Weissman were brave enough to walk a scientific path that very well could have ended in a dead end, and for that, they absolutely deserve their 2023 Nobel Prize.
This article originally appeared on Big Think, home of the brightest minds and biggest ideas of all time.

Scientists turn pee into power in Uganda
With conventional fuel cells as their model, researchers learned to use similar chemical reactions to make a fuel from microbes in pee.
At the edge of a dirt road flanked by trees and green mountains outside the town of Kisoro, Uganda, sits the concrete building that houses Sesame Girls School, where girls aged 11 to 19 can live, learn and, at least for a while, safely use a toilet. In many developing regions, toileting at night is especially dangerous for children. Without electrical power for lighting, kids may fall into the deep pits of the latrines through broken or unsteady floorboards. Girls are sometimes assaulted by men who hide in the dark.
For the Sesame School girls, though, bright LED lights, connected to tiny gadgets, chased the fears away. They got to use new, clean toilets lit by the power of their own pee. Some girls even used the light provided by the latrines to study.
Urine, whether animal or human, is more than waste. It’s a cheap and abundant resource. Each day across the globe, 8.1 billion humans make 4 billion gallons of pee. Cows, pigs, deer, elephants and other animals add more. By spending money to get rid of it, we waste a renewable resource that can serve more than one purpose. Microorganisms that feed on nutrients in urine can be used in a microbial fuel cell that generates electricity – or "pee power," as the Sesame girls called it.
Plus, urine contains water, phosphorus, potassium and nitrogen, the key ingredients plants need to grow and survive. Human urine could replace about 25 percent of current nitrogen and phosphorous fertilizers worldwide and could save water for gardens and crops. The average U.S. resident flushes a toilet bowl containing only pee and paper about six to seven times a day, which adds up to about 3,500 gallons of water down per year. Plus cows in the U.S. produce 231 gallons of the stuff each year.
Pee power
A conventional fuel cell uses chemical reactions to produce energy, as electrons move from one electrode to another to power a lightbulb or phone. Ioannis Ieropoulos, a professor and chair of Environmental Engineering at the University of Southampton in England, realized the same type of reaction could be used to make a fuel from microbes in pee.
Bacterial species like Shewanella oneidensis and Pseudomonas aeruginosa can consume carbon and other nutrients in urine and pop out electrons as a result of their digestion. In a microbial fuel cell, one electrode is covered in microbes, immersed in urine and kept away from oxygen. Another electrode is in contact with oxygen. When the microbes feed on nutrients, they produce the electrons that flow through the circuit from one electrod to another to combine with oxygen on the other side. As long as the microbes have fresh pee to chomp on, electrons keep flowing. And after the microbes are done with the pee, it can be used as fertilizer.
These microbes are easily found in wastewater treatment plants, ponds, lakes, rivers or soil. Keeping them alive is the easy part, says Ieropoulos. Once the cells start producing stable power, his group sequences the microbes and keeps using them.
Like many promising technologies, scaling these devices for mass consumption won’t be easy, says Kevin Orner, a civil engineering professor at West Virginia University. But it’s moving in the right direction. Ieropoulos’s device has shrunk from the size of about three packs of cards to a large glue stick. It looks and works much like a AAA battery and produce about the same power. By itself, the device can barely power a light bulb, but when stacked together, they can do much more—just like photovoltaic cells in solar panels. His lab has produced 1760 fuel cells stacked together, and with manufacturing support, there’s no theoretical ceiling, he says.
Although pure urine produces the most power, Ieropoulos’s devices also work with the mixed liquids of the wastewater treatment plants, so they can be retrofit into urban wastewater utilities.
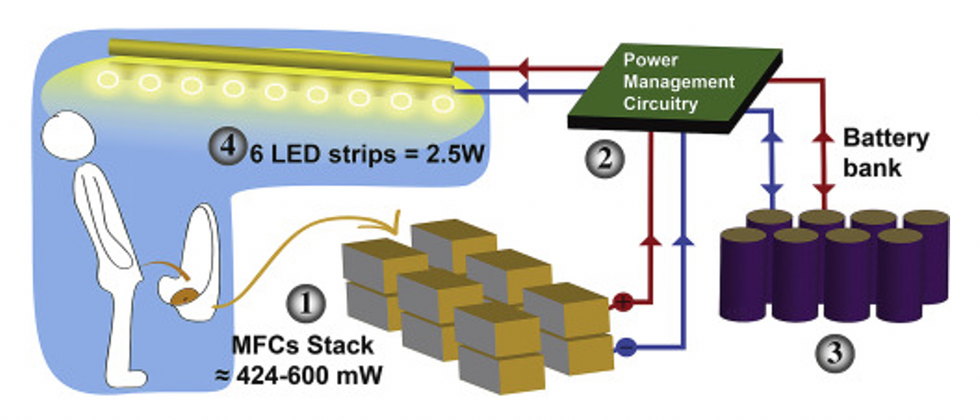
This image shows how the pee-powered system works. Pee feeds bacteria in the stack of fuel cells (1), which give off electrons (2) stored in parallel cylindrical cells (3). These cells are connected to a voltage regulator (4), which smooths out the electrical signal to ensure consistent power to the LED strips lighting the toilet.
Courtesy Ioannis Ieropoulos
Key to the long-term success of any urine reclamation effort, says Orner, is avoiding what he calls “parachute engineering”—when well-meaning scientists solve a problem with novel tech and then abandon it. “The way around that is to have either the need come from the community or to have an organization in a community that is committed to seeing a project operate and maintained,” he says.
Success with urine reclamation also depends on the economy. “If energy prices are low, it may not make sense to recover energy,” says Orner. “But right now, fertilizer prices worldwide are generally pretty high, so it may make sense to recover fertilizer and nutrients.” There are obstacles, too, such as few incentives for builders to incorporate urine recycling into new construction. And any hiccups like leaks or waste seepage will cost builders money and reputation. Right now, Orner says, the risks are just too high.
Despite the challenges, Ieropoulos envisions a future in which urine is passed through microbial fuel cells at wastewater treatment plants, retrofitted septic tanks, and building basements, and is then delivered to businesses to use as agricultural fertilizers. Although pure urine produces the most power, Ieropoulos’s devices also work with the mixed liquids of the wastewater treatment plants, so they can be retrofitted into urban wastewater utilities where they can make electricity from the effluent. And unlike solar cells, which are a common target of theft in some areas, nobody wants to steal a bunch of pee.
When Ieropoulos’s team returned to wrap up their pilot project 18 months later, the school’s director begged them to leave the fuel cells in place—because they made a major difference in students’ lives. “We replaced it with a substantial photovoltaic panel,” says Ieropoulos, They couldn’t leave the units forever, he explained, because of intellectual property reasons—their funders worried about theft of both the technology and the idea. But the photovoltaic replacement could be stolen, too, leaving the girls in the dark.
The story repeated itself at another school, in Nairobi, Kenya, as well as in an informal settlement in Durban, South Africa. Each time, Ieropoulos vowed to return. Though the pandemic has delayed his promise, he is resolute about continuing his work—it is a moral and legal obligation. “We've made a commitment to ourselves and to the pupils,” he says. “That's why we need to go back.”
Urine as fertilizer
Modern day industrial systems perpetuate the broken cycle of nutrients. When plants grow, they use up nutrients the soil. We eat the plans and excrete some of the nutrients we pass them into rivers and oceans. As a result, farmers must keep fertilizing the fields while our waste keeps fertilizing the waterways, where the algae, overfertilized with nitrogen, phosphorous and other nutrients grows out of control, sucking up oxygen that other marine species need to live. Few global communities remain untouched by the related challenges this broken chain create: insufficient clean water, food, and energy, and too much human and animal waste.
The Rich Earth Institute in Vermont runs a community-wide urine nutrient recovery program, which collects urine from homes and businesses, transports it for processing, and then supplies it as fertilizer to local farms.
One solution to this broken cycle is reclaiming urine and returning it back to the land. The Rich Earth Institute in Vermont is one of several organizations around the world working to divert and save urine for agricultural use. “The urine produced by an adult in one day contains enough fertilizer to grow all the wheat in one loaf of bread,” states their website.
Notably, while urine is not entirely sterile, it tends to harbor fewer pathogens than feces. That’s largely because urine has less organic matter and therefore less food for pathogens to feed on, but also because the urinary tract and the bladder have built-in antimicrobial defenses that kill many germs. In fact, the Rich Earth Institute says it’s safe to put your own urine onto crops grown for home consumption. Nonetheless, you’ll want to dilute it first because pee usually has too much nitrogen and can cause “fertilizer burn” if applied straight without dilution. Other projects to turn urine into fertilizer are in progress in Niger, South Africa, Kenya, Ethiopia, Sweden, Switzerland, The Netherlands, Australia, and France.
Eleven years ago, the Institute started a program that collects urine from homes and businesses, transports it for processing, and then supplies it as fertilizer to local farms. By 2021, the program included 180 donors producing over 12,000 gallons of urine each year. This urine is helping to fertilize hay fields at four partnering farms. Orner, the West Virginia professor, sees it as a success story. “They've shown how you can do this right--implementing it at a community level scale."

