Based on recent research, new therapies could promote a mix of viruses in the intestines to help prevent diseases of aging.
Story by Big Think
Our gut microbiome plays a substantial role in our health and well-being. Most research, however, focuses on bacteria, rather than the viruses that hide within them. Now, research from the University of Copenhagen, newly published in Nature Microbiology, found that people who live past age 100 have a greater diversity of bacteria-infecting viruses in their intestines than younger people. Furthermore, they found that the viruses are linked to changes in bacterial metabolism that may support mucosal integrity and resistance to pathogens.
The microbiota and aging
In the early 1970s, scientists discovered that the composition of our gut microbiota changes as we age. Recent studies have found that the changes are remarkably predictable and follow a pattern: The microbiota undergoes rapid, dramatic changes as toddlers transition to solid foods; further changes become less dramatic during childhood as the microbiota strikes a balance between the host and the environment; and as that balance is achieved, the microbiota remains mostly stable during our adult years (ages 18-60). However, that stability is lost as we enter our elderly years, and the microbiome undergoes dramatic reorganization. This discovery led scientists to question what causes this change and what effect it has on health.
Centenarians have a distinct gut community enriched in microorganisms that synthesize potent antimicrobial molecules that can kill multidrug-resistant pathogens.
“We are always eager to find out why some people live extremely long lives. Previous research has shown that the intestinal bacteria of old Japanese citizens produce brand-new molecules that make them resistant to pathogenic — that is, disease-promoting — microorganisms. And if their intestines are better protected against infection, well, then that is probably one of the things that cause them to live longer than others,” said Joachim Johansen, a researcher at the University of Copenhagen.
In 2021, a team of Japanese scientists set out to characterize the effect of this change on older people’s health. They specifically wanted to determine if people who lived to be over 100 years old — that is, centenarians — underwent changes that provided them with unique benefits. They discovered centenarians have a distinct gut community enriched in microorganisms that synthesize potent antimicrobial molecules that can kill multidrug-resistant pathogens, including Clostridioides difficile and Enterococcus faecium. In other words, the late-life shift in microbiota reduces an older person’s susceptibility to common gut pathogens.
Viruses can change alter the genes of bacteria
Although the late-in-life microbiota change could be beneficial to health, it remained unclear what facilitated this shift. To solve this mystery, Johansen and his colleagues turned their attention to an often overlooked member of the microbiome: viruses. “Our intestines contain billions of viruses living inside bacteria, and they could not care less about human cells; instead, they infect the bacterial cells. And seeing as there are hundreds of different types of bacteria in our intestines, there are also lots of bacterial viruses,” said Simon Rasmussen, Johansen’s research advisor.
Centenarians had a more diverse virome, including previously undescribed viral genera.
For decades, scientists have explored the possibility of phage therapy — that is, using viruses that infect bacteria (called bacteriophages or simply phages) to kill pathogens. However, bacteriophages can also enhance the bacteria they infect. For example, they can provide genes that help their bacterial host attack other bacteria or provide new metabolic capabilities. Both of these can change which bacteria colonize the gut and, in turn, protect against certain disease states.
Intestinal viruses give bacteria new abilities
Johansen and his colleagues were interested in what types of viruses centenarians had in their gut and whether those viruses carried genes that altered metabolism. They compared fecal samples of healthy centenarians (100+ year-olds) with samples from younger patients (18-100 year-olds). They found that the centenarians had a more diverse virome, including previously undescribed viral genera.
They also revealed an enrichment of genes supporting key steps in the sulfate metabolic pathway. The authors speculate that this translates to increased levels of microbially derived sulfide, which may lead to health-promoting outcomes, such as supporting mucosal integrity and resistance to potential pathogens.
“We have learned that if a virus pays a bacterium a visit, it may actually strengthen the bacterium. The viruses we found in the healthy Japanese centenarians contained extra genes that could boost the bacteria,” said Johansen.
Simon Rasmussen added, “If you discover bacteria and viruses that have a positive effect on the human intestinal flora, the obvious next step is to find out whether only some or all of us have them. If we are able to get these bacteria and their viruses to move in with the people who do not have them, more people could benefit from them.”
This article originally appeared on Big Think, home of the brightest minds and biggest ideas of all time.
Sign up for Big Think’s newsletter

A company has slashed the cost of assessing a person's genome to just $100. With lower costs - and as other genetic tools mature and evolve - a wave of new therapies could be coming in the near future.
In May 2022, Californian biotech Ultima Genomics announced that its UG 100 platform was capable of sequencing an entire human genome for just $100, a landmark moment in the history of the field. The announcement was particularly remarkable because few had previously heard of the company, a relative unknown in an industry long dominated by global giant Illumina which controls about 80 percent of the world’s sequencing market.
Ultima’s secret was to completely revamp many technical aspects of the way Illumina have traditionally deciphered DNA. The process usually involves first splitting the double helix DNA structure into single strands, then breaking these strands into short fragments which are laid out on a glass surface called a flow cell. When this flow cell is loaded into the sequencing machine, color-coded tags are attached to each individual base letter. A laser scans the bases individually while a camera simultaneously records the color associated with them, a process which is repeated until every single fragment has been sequenced.
Instead, Ultima has found a series of shortcuts to slash the cost and boost efficiency. “Ultima Genomics has developed a fundamentally new sequencing architecture designed to scale beyond conventional approaches,” says Josh Lauer, Ultima’s chief commercial officer.
This ‘new architecture’ is a series of subtle but highly impactful tweaks to the sequencing process ranging from replacing the costly flow cell with a silicon wafer which is both cheaper and allows more DNA to be read at once, to utilizing machine learning to convert optical data into usable information.
To put $100 genome in perspective, back in 2012 the cost of sequencing a single genome was around $10,000, a price tag which dropped to $1,000 a few years later. Before Ultima’s announcement, the cost of sequencing an individual genome was around $600.
Several studies have found that nearly 12 percent of healthy people who have their genome sequenced, then discover they have a variant pointing to a heightened risk of developing a disease that can be monitored, treated or prevented.
While Ultima’s new machine is not widely available yet, Illumina’s response has been rapid. In September 2022, the company unveiled the NovaSeq X series, which it describes as its fastest most cost-efficient sequencing platform yet, capable of sequencing genomes at $200, with further price cuts likely to follow.
But what will the rapidly tumbling cost of sequencing actually mean for medicine? “Well to start with, obviously it’s going to mean more people getting their genome sequenced,” says Michael Snyder, professor of genetics at Stanford University. “It'll be a lot more accessible to people.”
At the moment sequencing is mainly limited to certain cancer patients where it is used to inform treatment options, and individuals with undiagnosed illnesses. In the past, initiatives such as SeqFirst have attempted further widen access to genome sequencing based on growing amounts of research illustrating the potential benefits of the technology in healthcare. Several studies have found that nearly 12 percent of healthy people who have their genome sequenced, then discover they have a variant pointing to a heightened risk of developing a disease that can be monitored, treated or prevented.
“While whole genome sequencing is not yet widely used in the U.S., it has started to come into pediatric critical care settings such as newborn intensive care units,” says Professor Michael Bamshad, who heads the genetic medicine division in the University of Washington’s pediatrics department. “It is also being used more often in outpatient clinical genetics services, particularly when conventional testing fails to identify explanatory variants.”
But the cost of sequencing itself is only one part of the price tag. The subsequent clinical interpretation and genetic counselling services often come to several thousand dollars, a cost which insurers are not always willing to pay.
As a result, while Bamshad and others hope that the arrival of the $100 genome will create new opportunities to use genetic testing in innovative ways, the most immediate benefits are likely to come in the realm of research.
Bigger Data
There are numerous ways in which cheaper sequencing is likely to advance scientific research, for example the ability to collect data on much larger patient groups. This will be a major boon to scientists working on complex heterogeneous diseases such as schizophrenia or depression where there are many genes involved which all exert subtle effects, as well as substantial variance across the patient population. Bigger studies could help scientists identify subgroups of patients where the disease appears to be driven by similar gene variants, who can then be more precisely targeted with specific drugs.
If insurers can figure out the economics, Snyder even foresees a future where at a certain age, all of us can qualify for annual sequencing of our blood cells to search for early signs of cancer or the potential onset of other diseases like type 2 diabetes.
David Curtis, a genetics professor at University College London, says that scientists studying these illnesses have previously been forced to rely on genome-wide association studies which are limited because they only identify common gene variants. “We might see a significant increase in the number of large association studies using sequence data,” he says. “It would be far preferable to use this because it provides information about rare, potentially functional variants.”
Cheaper sequencing will also aid researchers working on diseases which have traditionally been underfunded. Bamshad cites cystic fibrosis, a condition which affects around 40,000 children and adults in the U.S., as one particularly pertinent example.
“Funds for gene discovery for rare diseases are very limited,” he says. “We’re one of three sites that did whole genome sequencing on 5,500 people with cystic fibrosis, but our statistical power is limited. A $100 genome would make it much more feasible to sequence everyone in the U.S. with cystic fibrosis and make it more likely that we discover novel risk factors and pathways influencing clinical outcomes.”
For progressive diseases that are more common like cancer and type 2 diabetes, as well as neurodegenerative conditions like multiple sclerosis and ALS, geneticists will be able to go even further and afford to sequence individual tumor cells or neurons at different time points. This will enable them to analyze how individual DNA modifications like methylation, change as the disease develops.
In the case of cancer, this could help scientists understand how tumors evolve to evade treatments. Within in a clinical setting, the ability to sequence not just one, but many different cells across a patient’s tumor could point to the combination of treatments which offer the best chance of eradicating the entire cancer.
“What happens at the moment with a solid tumor is you treat with one drug, and maybe 80 percent of that tumor is susceptible to that drug,” says Neil Ward, vice president and general manager in the EMEA region for genomics company PacBio. “But the other 20 percent of the tumor has already got mutations that make it resistant, which is probably why a lot of modern therapies extend life for sadly only a matter of months rather than curing, because they treat a big percentage of the tumor, but not the whole thing. So going forwards, I think that we will see genomics play a huge role in cancer treatments, through using multiple modalities to treat someone's cancer.”
If insurers can figure out the economics, Snyder even foresees a future where at a certain age, all of us can qualify for annual sequencing of our blood cells to search for early signs of cancer or the potential onset of other diseases like type 2 diabetes.
“There are companies already working on looking for cancer signatures in methylated DNA,” he says. “If it was determined that you had early stage cancer, pre-symptomatically, that could then be validated with targeted MRI, followed by surgery or chemotherapy. It makes a big difference catching cancer early. If there were signs of type 2 diabetes, you could start taking steps to mitigate your glucose rise, and possibly prevent it or at least delay the onset.”
This would already revolutionize the way we seek to prevent a whole range of illnesses, but others feel that the $100 genome could also usher in even more powerful and controversial preventative medicine schemes.
Newborn screening
In the eyes of Kári Stefánsson, the Icelandic neurologist who been a visionary for so many advances in the field of human genetics over the last 25 years, the falling cost of sequencing means it will be feasible to sequence the genomes of every baby born.
“We have recently done an analysis of genomes in Iceland and the UK Biobank, and in 4 percent of people you find mutations that lead to serious disease, that can be prevented or dealt with,” says Stefansson, CEO of deCODE genetics, a subsidiary of the pharmaceutical company Amgen. “This could transform our healthcare systems.”
As well as identifying newborns with rare diseases, this kind of genomic information could be used to compute a person’s risk score for developing chronic illnesses later in life. If for example, they have a higher than average risk of colon or breast cancer, they could be pre-emptively scheduled for annual colonoscopies or mammograms as soon as they hit adulthood.
To a limited extent, this is already happening. In the UK, Genomics England has launched the Newborn Genomes Programme, which plans to undertake whole-genome sequencing of up to 200,000 newborn babies, with the aim of enabling the early identification of rare genetic diseases.
"I have not had my own genome sequenced and I would not have wanted my parents to have agreed to this," Curtis says. "I don’t see that sequencing children for the sake of some vague, ill-defined benefits could ever be justifiable.”
However, some scientists feel that it is tricky to justify sequencing the genomes of apparently healthy babies, given the data privacy issues involved. They point out that we still know too little about the links which can be drawn between genetic information at birth, and risk of chronic illness later in life.
“I think there are very difficult ethical issues involved in sequencing children if there are no clear and immediate clinical benefits,” says Curtis. “They cannot consent to this process. I have not had my own genome sequenced and I would not have wanted my parents to have agreed to this. I don’t see that sequencing children for the sake of some vague, ill-defined benefits could ever be justifiable.”
Curtis points out that there are many inherent risks about this data being available. It may fall into the hands of insurance companies, and it could even be used by governments for surveillance purposes.
“Genetic sequence data is very useful indeed for forensic purposes. Its full potential has yet to be realized but identifying rare variants could provide a quick and easy way to find relatives of a perpetrator,” he says. “If large numbers of people had been sequenced in a healthcare system then it could be difficult for a future government to resist the temptation to use this as a resource to investigate serious crimes.”
While sequencing becoming more widely available will present difficult ethical and moral challenges, it will offer many benefits for society as a whole. Cheaper sequencing will help boost the diversity of genomic datasets which have traditionally been skewed towards individuals of white, European descent, meaning that much of the actionable medical information which has come out of these studies is not relevant to people of other ethnicities.
Ward predicts that in the coming years, the growing amount of genetic information will ultimately change the outcomes for many with rare, previously incurable illnesses.
“If you're the parent of a child that has a susceptible or a suspected rare genetic disease, their genome will get sequenced, and while sadly that doesn’t always lead to treatments, it’s building up a knowledge base so companies can spring up and target that niche of a disease,” he says. “As a result there’s a whole tidal wave of new therapies that are going to come to market over the next five years, as the genetic tools we have, mature and evolve.”
This article was first published by Leaps.org in October 2022.
Jamie Rettinger with his now fiance Amie Purnel-Davis, who helped him through the clinical trial.
Jamie Rettinger was still in his thirties when he first noticed a tiny streak of brown running through the thumbnail of his right hand. It slowly grew wider and the skin underneath began to deteriorate before he went to a local dermatologist in 2013. The doctor thought it was a wart and tried scooping it out, treating the affected area for three years before finally removing the nail bed and sending it off to a pathology lab for analysis.
"I have some bad news for you; what we removed was a five-millimeter melanoma, a cancerous tumor that often spreads," Jamie recalls being told on his return visit. "I'd never heard of cancer coming through a thumbnail," he says. None of his doctors had ever mentioned it either. "I just thought I was being treated for a wart." But nothing was healing and it continued to bleed.
A few months later a surgeon amputated the top half of his thumb. Lymph node biopsy tested negative for spread of the cancer and when the bandages finally came off, Jamie thought his medical issues were resolved.
Melanoma is the deadliest form of skin cancer. About 85,000 people are diagnosed with it each year in the U.S. and more than 8,000 die of the cancer when it spreads to other parts of the body, according to the Centers for Disease Control and Prevention (CDC).
There are two peaks in diagnosis of melanoma; one is in younger women ages 30-40 and often is tied to past use of tanning beds; the second is older men 60+ and is related to outdoor activity from farming to sports. Light-skinned people have a twenty-times greater risk of melanoma than do people with dark skin.
"When I graduated from medical school, in 2005, melanoma was a death sentence" --Diwakar Davar.
Jamie had a follow up PET scan about six months after his surgery. A suspicious spot on his lung led to a biopsy that came back positive for melanoma. The cancer had spread. Treatment with a monoclonal antibody (nivolumab/Opdivo®) didn't prove effective and he was referred to the UPMC Hillman Cancer Center in Pittsburgh, a four-hour drive from his home in western Ohio.
An alternative monoclonal antibody treatment brought on such bad side effects, diarrhea as often as 15 times a day, that it took more than a week of hospitalization to stabilize his condition. The only options left were experimental approaches in clinical trials.
Early research
"When I graduated from medical school, in 2005, melanoma was a death sentence" with a cure rate in the single digits, says Diwakar Davar, 39, an oncologist at UPMC Hillman Cancer Center who specializes in skin cancer. That began to change in 2010 with introduction of the first immunotherapies, monoclonal antibodies, to treat cancer. The antibodies attach to PD-1, a receptor on the surface of T cells of the immune system and on cancer cells. Antibody treatment boosted the melanoma cure rate to about 30 percent. The search was on to understand why some people responded to these drugs and others did not.
At the same time, there was a growing understanding of the role that bacteria in the gut, the gut microbiome, plays in helping to train and maintain the function of the body's various immune cells. Perhaps the bacteria also plays a role in shaping the immune response to cancer therapy.
One clue came from genetically identical mice. Animals ordered from different suppliers sometimes responded differently to the experiments being performed. That difference was traced to different compositions of their gut microbiome; transferring the microbiome from one animal to another in a process known as fecal transplant (FMT) could change their responses to disease or treatment.
When researchers looked at humans, they found that the patients who responded well to immunotherapies had a gut microbiome that looked like healthy normal folks, but patients who didn't respond had missing or reduced strains of bacteria.
Davar and his team knew that FMT had a very successful cure rate in treating the gut dysbiosis of Clostridioides difficile, a persistant intestinal infection, and they wondered if a fecal transplant from a patient who had responded well to cancer immunotherapy treatment might improve the cure rate of patients who did not originally respond to immunotherapies for melanoma.
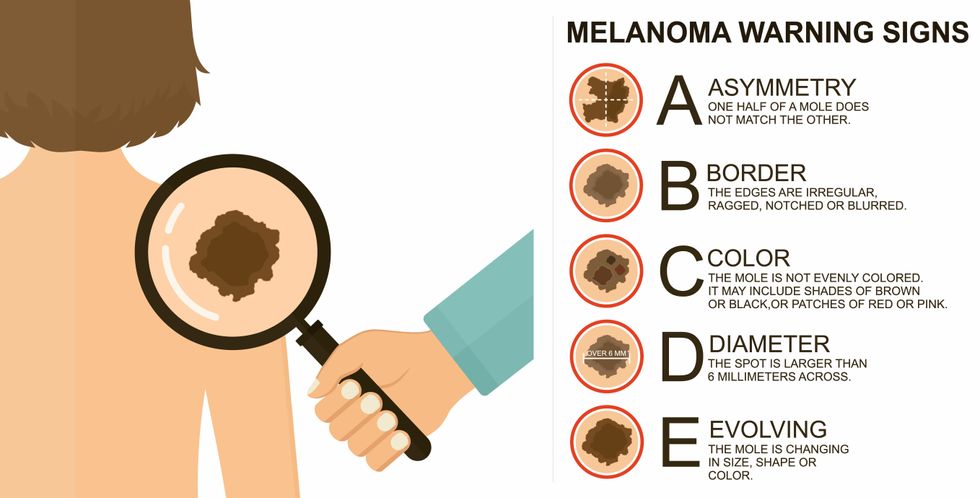
The ABCDE of melanoma detection
Adobe Stock
Clinical trial
"It was pretty weird, I was totally blasted away. Who had thought of this?" Jamie first thought when the hypothesis was explained to him. But Davar's explanation that the procedure might restore some of the beneficial bacterial his gut was lacking, convinced him to try. He quickly signed on in October 2018 to be the first person in the clinical trial.
Fecal donations go through the same safety procedures of screening for and inactivating diseases that are used in processing blood donations to make them safe for transfusion. The procedure itself uses a standard hollow colonoscope designed to screen for colon cancer and remove polyps. The transplant is inserted through the center of the flexible tube.
Most patients are sedated for procedures that use a colonoscope but Jamie doesn't respond to those drugs: "You can't knock me out. I was watching them on the TV going up my own butt. It was kind of unreal at that point," he says. "There were about twelve people in there watching because no one had seen this done before."
A test two weeks after the procedure showed that the FMT had engrafted and the once-missing bacteria were thriving in his gut. More importantly, his body was responding to another monoclonal antibody (pembrolizumab/Keytruda®) and signs of melanoma began to shrink. Every three months he made the four-hour drive from home to Pittsburgh for six rounds of treatment with the antibody drug.
"We were very, very lucky that the first patient had a great response," says Davar. "It allowed us to believe that even though we failed with the next six, we were on the right track. We just needed to tweak the [fecal] cocktail a little better" and enroll patients in the study who had less aggressive tumor growth and were likely to live long enough to complete the extensive rounds of therapy. Six of 15 patients responded positively in the pilot clinical trial that was published in the journal Science.
Davar believes they are beginning to understand the biological mechanisms of why some patients initially do not respond to immunotherapy but later can with a FMT. It is tied to the background level of inflammation produced by the interaction between the microbiome and the immune system. That paper is not yet published.
Surviving cancer
It has been almost a year since the last in his series of cancer treatments and Jamie has no measurable disease. He is cautiously optimistic that his cancer is not simply in remission but is gone for good. "I'm still scared every time I get my scans, because you don't know whether it is going to come back or not. And to realize that it is something that is totally out of my control."
"It was hard for me to regain trust" after being misdiagnosed and mistreated by several doctors he says. But his experience at Hillman helped to restore that trust "because they were interested in me, not just fixing the problem."
He is grateful for the support provided by family and friends over the last eight years. After a pause and a sigh, the ruggedly built 47-year-old says, "If everyone else was dead in my family, I probably wouldn't have been able to do it."
"I never hesitated to ask a question and I never hesitated to get a second opinion." But Jamie acknowledges the experience has made him more aware of the need for regular preventive medical care and a primary care physician. That person might have caught his melanoma at an earlier stage when it was easier to treat.
Davar continues to work on clinical studies to optimize this treatment approach. Perhaps down the road, screening the microbiome will be standard for melanoma and other cancers prior to using immunotherapies, and the FMT will be as simple as swallowing a handful of freeze-dried capsules off the shelf rather than through a colonoscopy. Earlier this year, the Food and Drug Administration approved the first oral fecal microbiota product for C. difficile, hopefully paving the way for more.
An older version of this hit article was first published on May 18, 2021


