Is Finding Out Your Baby’s Genetics A New Responsibility of Parenting?
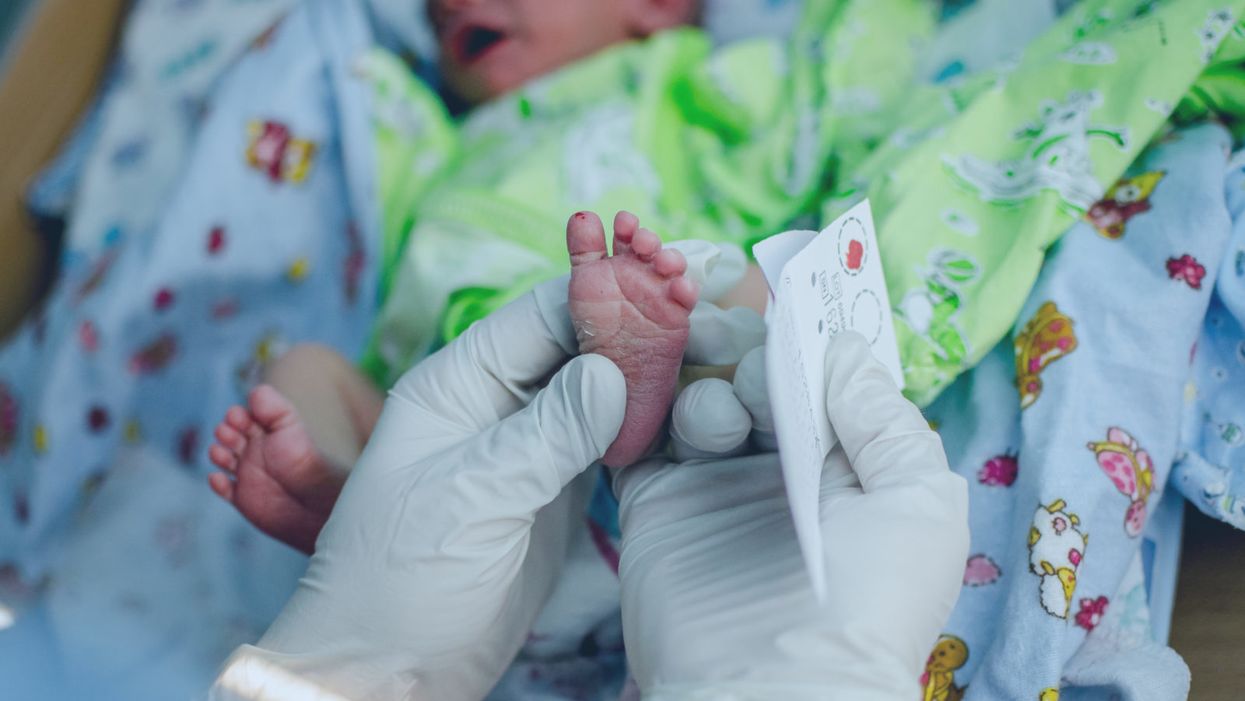
A doctor pricks the heel of a newborn for a blood test.
Hours after a baby is born, its heel is pricked with a lancet. Drops of the infant's blood are collected on a porous card, which is then mailed to a state laboratory. The dried blood spots are screened for around thirty conditions, including phenylketonuria (PKU), the metabolic disorder that kick-started this kind of newborn screening over 60 years ago. In the U.S., parents are not asked for permission to screen their child. Newborn screening programs are public health programs, and the assumption is that no good parent would refuse a screening test that could identify a serious yet treatable condition in their baby.
Learning as much as you can about your child's health might seem like a natural obligation of parenting. But it's an assumption that I think needs to be much more closely examined.
Today, with the introduction of genome sequencing into clinical medicine, some are asking whether newborn screening goes far enough. As the cost of sequencing falls, should parents take a more expansive look at their children's health, learning not just whether they have a rare but treatable childhood condition, but also whether they are at risk for untreatable conditions or for diseases that, if they occur at all, will strike only in adulthood? Should genome sequencing be a part of every newborn's care?
It's an idea that appeals to Anne Wojcicki, the founder and CEO of the direct-to-consumer genetic testing company 23andMe, who in a 2016 interview with The Guardian newspaper predicted that having newborns tested would soon be considered standard practice—"as critical as testing your cholesterol"—and a new responsibility of parenting. Wojcicki isn't the only one excited to see everyone's genes examined at birth. Francis Collins, director of the National Institutes of Health and perhaps the most prominent advocate of genomics in the United States, has written that he is "almost certain … that whole-genome sequencing will become part of new-born screening in the next few years." Whether that would happen through state-mandated screening programs, or as part of routine pediatric care—or perhaps as a direct-to-consumer service that parents purchase at birth or receive as a baby-shower gift—is not clear.
Learning as much as you can about your child's health might seem like a natural obligation of parenting. But it's an assumption that I think needs to be much more closely examined, both because the results that genome sequencing can return are more complex and more uncertain than one might expect, and because parents are not actually responsible for their child's lifelong health and well-being.
What is a parent supposed to do about such a risk except worry?
Existing newborn screening tests look for the presence of rare conditions that, if identified early in life, before the child shows any symptoms, can be effectively treated. Sequencing could identify many of these same kinds of conditions (and it might be a good tool if it could be targeted to those conditions alone), but it would also identify gene variants that confer an increased risk rather than a certainty of disease. Occasionally that increased risk will be significant. About 12 percent of women in the general population will develop breast cancer during their lives, while those who have a harmful BRCA1 or BRCA2 gene variant have around a 70 percent chance of developing the disease. But for many—perhaps most—conditions, the increased risk associated with a particular gene variant will be very small. Researchers have identified over 600 genes that appear to be associated with schizophrenia, for example, but any one of those confers only a tiny increase in risk for the disorder. What is a parent supposed to do about such a risk except worry?
Sequencing results are uncertain in other important ways as well. While we now have the ability to map the genome—to create a read-out of the pairs of genetic letters that make up a person's DNA—we are still learning what most of it means for a person's health and well-being. Researchers even have a name for gene variants they think might be associated with a disease or disorder, but for which they don't have enough evidence to be sure. They are called "variants of unknown (or uncertain) significance (VUS), and they pop up in most people's sequencing results. In cancer genetics, where much research has been done, about 1 in 5 gene variants are reclassified over time. Most are downgraded, which means that a good number of VUS are eventually designated benign.
While one parent might reasonably decide to learn about their child's risk for a condition about which nothing can be done medically, a different, yet still thoroughly reasonable, parent might prefer to remain ignorant so that they can enjoy the time before their child is afflicted.
Then there's the puzzle of what to do about results that show increased risk or even certainty for a condition that we have no idea how to prevent. Some genomics advocates argue that even if a result is not "medically actionable," it might have "personal utility" because it allows parents to plan for their child's future needs, to enroll them in research, or to connect with other families whose children carry the same genetic marker.
Finding a certain gene variant in one child might inform parents' decisions about whether to have another—and if they do, about whether to use reproductive technologies or prenatal testing to select against that variant in a future child. I have no doubt that for some parents these personal utility arguments are persuasive, but notice how far we've now strayed from the serious yet treatable conditions that motivated governments to set up newborn screening programs, and to mandate such testing for all.
Which brings me to the other problem with the call for sequencing newborn babies: the idea that even if it's not what the law requires, it's what good parents should do. That idea is very compelling when we're talking about sequencing results that show a serious threat to the child's health, especially when interventions are available to prevent or treat that condition. But as I have shown, many sequencing results are not of this type.
While one parent might reasonably decide to learn about their child's risk for a condition about which nothing can be done medically, a different, yet still thoroughly reasonable, parent might prefer to remain ignorant so that they can enjoy the time before their child is afflicted. This parent might decide that the worry—and the hypervigilence it could inspire in them—is not in their child's best interest, or indeed in their own. This parent might also think that it should be up to the child, when he or she is older, to decide whether to learn about his or her risk for adult-onset conditions, especially given that many adults at high familial risk for conditions like Alzheimer's or Huntington's disease choose never to be tested. This parent will value the child's future autonomy and right not to know more than they value the chance to prepare for a health risk that won't strike the child until 40 or 50 years in the future.
Parents are not obligated to learn about their children's risk for a condition that cannot be prevented, has a small risk of occurring, or that would appear only in adulthood.
Contemporary understandings of parenting are famously demanding. We are asked to do everything within our power to advance our children's health and well-being—to act always in our children's best interests. Against that backdrop, the need to sequence every newborn baby's genome might seem obvious. But we should be skeptical. Many sequencing results are complex and uncertain. Parents are not obligated to learn about their children's risk for a condition that cannot be prevented, has a small risk of occurring, or that would appear only in adulthood. To suggest otherwise is to stretch parental responsibilities beyond the realm of childhood and beyond factors that parents can control.
Can Biotechnology Take the Allergies Out of Cats?
From a special food to a vaccine and gene editing, new technologies may offer solutions for cat lovers with allergies.
Amy Bitterman, who teaches at Rutgers Law School in Newark, gets enormous pleasure from her three mixed-breed rescue cats, Spike, Dee, and Lucy. To manage her chronically stuffy nose, three times a week she takes Allegra D, which combines the antihistamine fexofenadine with the decongestant pseudoephedrine. Amy's dog allergy is rougher--so severe that when her sister launched a business, Pet Care By Susan, from their home in Edison, New Jersey, they knew Susan would have to move elsewhere before she could board dogs. Amy has tried to visit their brother, who owns a Labrador Retriever, taking Allegra D beforehand. But she began sneezing, and then developed watery eyes and phlegm in her chest.
"It gets harder and harder to breathe," she says.
Animal lovers have long dreamed of "hypo-allergenic" cats and dogs. Although to date, there is no such thing, biotechnology is beginning to provide solutions for cat-lovers. Cats are a simpler challenge than dogs. Dog allergies involve as many as seven proteins. But up to 95 percent of people who have cat allergies--estimated at 10 to 30 percent of the population in North America and Europe--react to one protein, Fel d1. Interestingly, cats don't seem to need Fel d1. There are cats who don't produce much Fel d1 and have no known health problems.
The current technologies fight Fel d1 in ingenious ways. Nestle Purina reached the market first with a cat food, Pro Plan LiveClear, launched in the U.S. a year and a half ago. It contains Fel d1 antibodies from eggs that in effect neutralize the protein. HypoCat, a vaccine for cats, induces them to create neutralizing antibodies to their own Fel d1. It may be available in the United States by 2024, says Gary Jennings, chief executive officer of Saiba Animal Health, a University of Zurich spin-off. Another approach, using the gene-editing tool CRISPR to create a medication that would splice out Fel d1 genes in particular tissues, is the furthest from fruition.
"Our goal was to ensure that whatever we do has no negative impact on the cat."
Customer demand is high. "We already have a steady stream of allergic cat owners contacting us desperate to have access to the vaccine or participate in the testing program," Jennings said. "There is a major unmet medical need."
More than a third of Americans own a cat (while half own a dog), and pet ownership is rising. With more Americans living alone, pets may be just the right amount of company. But the number of Americans with asthma increases every year. Of that group, some 20 to 30 percent have pet allergies that could trigger a possibly deadly attack. It is not clear how many pets end up in shelters because their owners could no longer manage allergies. Instead, allergists commonly report that their patients won't give up a beloved companion.
No one can completely avoid Fel d1, which clings to clothing and lands everywhere cat-owners go, even in schools and new homes never occupied by cats. Myths among cat-lovers may lead them to underestimate their own level of risk. Short hair doesn't help: the length of cat hair doesn't affect the production of Fel d1. Bathing your cat will likely upset it and accomplish little. Washing cuts the amount on its skin and fur only for two days. In one study, researchers measured the Fel d1 in the ambient air in a small chamber occupied by a cat—and then washed the cat. Three hours later, with the cat in the chamber again, the measurable Fel d1 in the air was lower. But this benefit was gone after 24 hours.
For years, the best option has been shots for people that prompt protective antibodies. Bitterman received dog and cat allergy injections twice a week as a child. However, these treatments require up to 100 injections over three to five years, and, as in her case, the effect may be partial or wear off. Even if you do opt for shots, treating the cat also makes sense, since you could protect more than one allergic member of your household and any allergic visitors as well.
An Allergy-Neutralizing Diet
Cats produce much of their Fel d1 in their saliva, which then spreads it to their fur when they groom, observed Nestle Purina immunologist Ebenezer Satyaraj. He realized that this made saliva—and therefore a cat's mouth--an unusually effective site for change. Hens exposed to Fel d1 produce their own antibodies, which survive in their eggs. The team coated LiveClear food with a powder form of these eggs; once in a cat's mouth, the chicken antibody binds to the Fel d1 in the cat's saliva, neutralizing it.
The results are partial: In a study with 105 cats, the level of active Fel d1 in their fur had dropped on average by 47 percent after ten weeks eating LiveClear. Cats that produced more Fel d1 at baseline had a more robust response, with a drop of up to 71 percent. A safety study found no effects on cats after six months on the diet. "Our goal was to ensure that whatever we do has no negative impact on the cat," Satyaraj said. Might a dogfood that minimizes dog allergens be on the way? "There is some early work," he said.
A Vaccine
This is a year when vaccines changed the lives of billions. Saiba's vaccine, HypoCat, delivers recombinant Fel d1 and the coat from a plant virus (the Cucumber mosaic virus) without any vital genetic information. The viral coat serves as a carrier. A cat would need shots once or twice a year to produce antibodies that neutralize Fel d1.
HypoCat works much like any vaccine, with the twist that the enemy is the cat's own protein. Is that safe? Saiba's team has followed 70 cats treated with the vaccine over two years and they remain healthy. Again the active Fel d1 doesn't disappear but diminishes. The team asked 10 people with cat allergies to report on their symptoms when they pet their vaccinated cats. Eight of them could pet their cat for nearly a half hour before their symptoms began, compared with an average of 17 minutes before the vaccine.
Jennings hopes to develop a HypoDog shot with a similar approach. However, the goal would be to target four or five proteins in one vaccine, and that increases the risk of hurting the dog. In the meantime, allergic dog-lovers considering an expensive breeder dog might think again: Independent research does not support the idea that any breed of dog produces less dander in the home. In fact, one well-designed study found that Spanish water dogs, Airedales, poodles and Labradoodles--breeds touted as hypo-allergenic--had significantly more of the most common allergen on their coat than an ordinary Lab and the control group.
Gene Editing
One day you might be able to bring your cat to the vet once a year for an injection that would modify specific tissues so they wouldn't produce Fel d1.
Nicole Brackett, a postdoctoral scientist at Viriginia-based Indoor Biotechnologies, which specializes in manufacturing biologics for allergy and asthma, most recently has used CRISPR to identify Fel d1 genetic sequences in cells from 50 domestic cats and 24 exotic ones. She learned that the sequences vary substantially from one cat to the next. This discovery, she says, backs up the observations that Fel d1 doesn't have a vital purpose.
The next step will be a CRISPR knockout of the relevant genes in cells from feline salivary glands, a prime source of Fel d1. Although the company is considering using CRISPR to edit the genes in a cat embryo and possibly produce a Fel d1-free cat, designer cats won't be its ultimate product. Instead, the company aims to produce injections that could treat any cat.
Reducing pet allergens at home could have a compound benefit, Indoor Biotechnologies founder Martin Chapman, an immunologist, notes: "When you dampen down the response to one allergen, you could also dampen it down to multiple allergens." As allergies become more common around the world, that's especially good news.
The large genetic studies underlying certain disease risk tests have primarily been done in populations of European ancestry, limiting their accuracy.
Earlier this year, California-based Ambry Genetics announced that it was discontinuing a test meant to estimate a person's risk of developing prostate or breast cancer. The test looks for variations in a person's DNA that are known to be associated with these cancers.
Known as a polygenic risk score, this type of test adds up the effects of variants in many genes — often in the dozens or hundreds — and calculates a person's risk of developing a particular health condition compared to other people. In this way, polygenic risk scores are different from traditional genetic tests that look for mutations in single genes, such as BRCA1 and BRCA2, which raise the risk of breast cancer.
Traditional genetic tests look for mutations that are relatively rare in the general population but have a large impact on a person's disease risk, like BRCA1 and BRCA2. By contrast, polygenic risk scores scan for more common genetic variants that, on their own, have a small effect on risk. Added together, however, they can raise a person's risk for developing disease.
These scores could become a part of routine healthcare in the next few years. Researchers are developing polygenic risk scores for cancer, heart, disease, diabetes and even depression. Before they can be rolled out widely, they'll have to overcome a key limitation: racial bias.
"The issue with these polygenic risk scores is that the scientific studies which they're based on have primarily been done in individuals of European ancestry," says Sara Riordan, president of the National Society of Genetics Counselors. These scores are calculated by comparing the genetic data of people with and without a particular disease. To make these scores accurate, researchers need genetic data from tens or hundreds of thousands of people.
Myriad's old test would have shown that a Black woman had twice as high of a risk for breast cancer compared to the average woman even if she was at low or average risk.
A 2018 analysis found that 78% of participants included in such large genetic studies, known as genome-wide association studies, were of European descent. That's a problem, because certain disease-associated genetic variants don't appear equally across different racial and ethnic groups. For example, a particular variant in the TTR gene, known as V1221, occurs more frequently in people of African descent. In recent years, the variant has been found in 3 to 4 percent of individuals of African ancestry in the United States. Mutations in this gene can cause protein to build up in the heart, leading to a higher risk of heart failure. A polygenic risk score for heart disease based on genetic data from mostly white people likely wouldn't give accurate risk information to African Americans.
Accuracy in genetic testing matters because such polygenic risk scores could help patients and their doctors make better decisions about their healthcare.
For instance, if a polygenic risk score determines that a woman is at higher-than-average risk of breast cancer, her doctor might recommend more frequent mammograms — X-rays that take a picture of the breast. Or, if a risk score reveals that a patient is more predisposed to heart attack, a doctor might prescribe preventive statins, a type of cholesterol-lowering drug.
"Let's be clear, these are not diagnostic tools," says Alicia Martin, a population and statistical geneticist at the Broad Institute of MIT and Harvard. "We can't use a polygenic score to say you will or will not get breast cancer or have a heart attack."
But combining a patient's polygenic risk score with other factors that affect disease risk — like age, weight, medication use or smoking status — may provide a better sense of how likely they are to develop a specific health condition than considering any one risk factor one its own. The accuracy of polygenic risk scores becomes even more important when considering that these scores may be used to guide medication prescription or help patients make decisions about preventive surgery, such as a mastectomy.
In a study published in September, researchers used results from large genetics studies of people with European ancestry and data from the UK Biobank to calculate polygenic risk scores for breast and prostate cancer for people with African, East Asian, European and South Asian ancestry. They found that they could identify individuals at higher risk of breast and prostate cancer when they scaled the risk scores within each group, but the authors say this is only a temporary solution. Recruiting more diverse participants for genetics studies will lead to better cancer detection and prevent, they conclude.
Recent efforts to do just that are expected to make these scores more accurate in the future. Until then, some genetics companies are struggling to overcome the European bias in their tests.
Acknowledging the limitations of its polygenic risk score, Ambry Genetics said in April that it would stop offering the test until it could be recalibrated. The company launched the test, known as AmbryScore, in 2018.
"After careful consideration, we have decided to discontinue AmbryScore to help reduce disparities in access to genetic testing and to stay aligned with current guidelines," the company said in an email to customers. "Due to limited data across ethnic populations, most polygenic risk scores, including AmbryScore, have not been validated for use in patients of diverse backgrounds." (The company did not make a spokesperson available for an interview for this story.)
In September 2020, the National Comprehensive Cancer Network updated its guidelines to advise against the use of polygenic risk scores in routine patient care because of "significant limitations in interpretation." The nonprofit, which represents 31 major cancer cancers across the United States, said such scores could continue to be used experimentally in clinical trials, however.
Holly Pederson, director of Medical Breast Services at the Cleveland Clinic, says the realization that polygenic risk scores may not be accurate for all races and ethnicities is relatively recent. Pederson worked with Salt Lake City-based Myriad Genetics, a leading provider of genetic tests, to improve the accuracy of its polygenic risk score for breast cancer.
The company announced in August that it had recalibrated the test, called RiskScore, for women of all ancestries. Previously, Myriad did not offer its polygenic risk score to women who self-reported any ancestry other than sole European or Ashkenazi ancestry.
"Black women, while they have a similar rate of breast cancer to white women, if not lower, had twice as high of a polygenic risk score because the development and validation of the model was done in white populations," Pederson said of the old test. In other words, Myriad's old test would have shown that a Black woman had twice as high of a risk for breast cancer compared to the average woman even if she was at low or average risk.
To develop and validate the new score, Pederson and other researchers assessed data from more than 275,000 women, including more than 31,000 African American women and nearly 50,000 women of East Asian descent. They looked at 56 different genetic variants associated with ancestry and 93 associated with breast cancer. Interestingly, they found that at least 95% of the breast cancer variants were similar amongst the different ancestries.
The company says the resulting test is now more accurate for all women across the board, but Pederson cautions that it's still slightly less accurate for Black women.
"It's not only the lack of data from Black women that leads to inaccuracies and a lack of validation in these types of risk models, it's also the pure genomic diversity of Africa," she says, noting that Africa is the most genetically diverse continent on the planet. "We just need more data, not only in American Black women but in African women to really further characterize that continent."
Martin says it's problematic that such scores are most accurate for white people because they could further exacerbate health disparities in traditionally underserved groups, such as Black Americans. "If we were to set up really representative massive genetic studies, we would do a much better job at predicting genetic risk for everybody," she says.
Earlier this year, the National Institutes of Health awarded $38 million to researchers to improve the accuracy of polygenic risk scores in diverse populations. Researchers will create new genome datasets and pool information from existing ones in an effort to diversify the data that polygenic scores rely on. They plan to make these datasets available to other scientists to use.
"By having adequate representation, we can ensure that the results of a genetic test are widely applicable," Riordan says.

