The New Prospective Parenthood: When Does More Info Become Too Much?
Obstetric ultrasound of a fourth-month fetus.
Peggy Clark was 12 weeks pregnant when she went in for a nuchal translucency (NT) scan to see whether her unborn son had Down syndrome. The sonographic scan measures how much fluid has accumulated at the back of the baby's neck: the more fluid, the higher the likelihood of an abnormality. The technician said the baby was in such an odd position, the test couldn't be done. Clark, whose name has been changed to protect her privacy, was told to come back in a week and a half to see if the baby had moved.
"With the growing sophistication of prenatal tests, it seems that the more questions are answered, the more new ones arise."
"It was like the baby was saying, 'I don't want you to know,'" she recently recalled.
When they went back, they found the baby had a thickened neck. It's just one factor in identifying Down's, but it's a strong indication. At that point, she was 13 weeks and four days pregnant. She went to the doctor the next day for a blood test. It took another two weeks for the results, which again came back positive, though there was still a .3% margin of error. Clark said she knew she wanted to terminate the pregnancy if the baby had Down's, but she didn't want the guilt of knowing there was a small chance the tests were wrong. At that point, she was too late to do a Chorionic villus sampling (CVS), when chorionic villi cells are removed from the placenta and sequenced. And she was too early to do an amniocentesis, which isn't done until between 14 and 20 weeks of the pregnancy. So she says she had to sit and wait, calling those few weeks "brutal."
By the time they did the amnio, she was already nearly 18 weeks pregnant and was getting really big. When that test also came back positive, she made the anguished decision to end the pregnancy.
Now, three years after Clark's painful experience, a newer form of prenatal testing routinely gives would-be parents more information much earlier on, especially for women who are over 35. As soon as nine weeks into their pregnancies, women can have a simple blood test to determine if there are abnormalities in the DNA of chromosomes 21, which indicates Down syndrome, as well as in chromosomes 13 and 18. Using next-generation sequencing technologies, the test separates out and examines circulating fetal cells in the mother's blood, which eliminates the risks of drawing fluid directly from the fetus or placenta.
"Finding out your baby has Down syndrome at 11 or 12 weeks is much easier for parents to make any decision they may want to make, as opposed to 16 or 17 weeks," said Dr. Leena Nathan, an obstetrician-gynecologist in UCLA's healthcare system. "People are much more willing or able to perhaps make a decision to terminate the pregnancy."
But with the growing sophistication of prenatal tests, it seems that the more questions are answered, the more new ones arise--questions that previous generations have never had to face. And as genomic sequencing improves in its predictive accuracy at the earliest stages of life, the challenges only stand to increase. Imagine, for example, learning your child's lifetime risk of breast cancer when you are ten weeks pregnant. Would you terminate if you knew she had a 70 percent risk? What about 40 percent? Lots of hard questions. Few easy answers. Once the cost of whole genome sequencing drops low enough, probably within the next five to ten years according to experts, such comprehensive testing may become the new standard of care. Welcome to the future of prospective parenthood.
"In one way, it's a blessing to have this information. On the other hand, it's very difficult to deal with."
How Did We Get Here?
Prenatal testing is not new. In 1979, amniocentesis was used to detect whether certain inherited diseases had been passed on to the fetus. Through the 1980s, parents could be tested to see if they carried disease like Tay-Sachs, Sickle cell anemia, Cystic fibrosis and Duchenne muscular dystrophy. By the early 1990s, doctors could test for even more genetic diseases and the CVS test was beginning to become available.
A few years later, a technique called preimplantation genetic diagnosis (PGD) emerged, in which embryos created in a lab with sperm and harvested eggs would be allowed to grow for several days and then cells would be removed and tested to see if any carried genetic diseases. Those that weren't affected could be transferred back to the mother. Once in vitro fertilization (IVF) took off, so did genetic testing. The labs test the embryonic cells and get them back to the IVF facilities within 24 hours so that embryo selection can occur. In the case of IVF, genetic tests are done so early, parents don't even have to decide whether to terminate a pregnancy. Embryos with issues often aren't even used.
"It was a very expensive endeavor but exciting to see our ability to avoid disorders, especially for families that don't want to terminate a pregnancy," said Sara Katsanis, an expert in genetic testing who teaches at Duke University. "In one way, it's a blessing to have this information (about genetic disorders). On the other hand, it's very difficult to deal with. To make that decision about whether to terminate a pregnancy is very hard."
Just Because We Can, Does It Mean We Should?
Parents in the future may not only find out whether their child has a genetic disease but will be able to potentially fix the problem through a highly controversial process called gene editing. But because we can, does it mean we should? So far, genes have been edited in other species, but to date, the procedure has not been used on an unborn child for reproductive purposes apart from research.
"There's a lot of bioethics debate and convening of groups to try to figure out where genetic manipulation is going to be useful and necessary, and where it is going to need some restrictions," said Katsanis. She notes that it's very useful in areas like cancer research, so one wouldn't want to over-regulate it.
There are already some criteria as to which genes can be manipulated and which should be left alone, said Evan Snyder, professor and director of the Center for Stem Cells and Regenerative Medicine at Sanford Children's Health Research Center in La Jolla, Calif. He noted that genes don't stand in isolation. That is, if you modify one that causes disease, will it disrupt others? There may be unintended consequences, he added.
"As the technical dilemmas get fixed, some of the ethical dilemmas get fixed. But others arise. It's kind of like ethical whack-a-mole."
But gene editing of embryos may take years to become an acceptable practice, if ever, so a more pressing issue concerns the rationale behind embryo selection during IVF. Prospective parents can end up with anywhere from zero to thirty embryos from the procedure and must choose only one (rarely two) to implant. Since embryos are routinely tested now for certain diseases, and selected or discarded based on that information, should it be ethical—and legal—to make selections based on particular traits, too? To date so far, parents can select for gender, but no other traits. Whether trait selection becomes routine is a matter of time and business opportunity, Katsanis said. So far, the old-fashioned way of making a baby combined with the luck of the draw seems to be the preferred method for the marketplace. But that could change.
"You can easily see a family deciding not to implant a lethal gene for Tay-Sachs or Duchene or Cystic fibrosis. It becomes more ethically challenging when you make a decision to implant girls and not any of the boys," said Snyder. "And then as we get better and better, we can start assigning genes to certain skills and this starts to become science fiction."
Once a pregnancy occurs, prospective parents of all stripes will face decisions about whether to keep the fetus based on the information that increasingly robust prenatal testing will provide. What influences their decision is the crux of another ethical knot, said Snyder. A clear-cut rationale would be if the baby is anencephalic, or it has no brain. A harder one might be, "It's a girl, and I wanted a boy," or "The child will only be 5' 2" tall in adulthood."
"Those are the extremes, but the ultimate question is: At what point is it a legitimate response to say, I don't want to keep this baby?'" he said. Of course, people's responses will vary, so the bigger conundrum for society is: Where should a line be drawn—if at all? Should a woman who is within the legal scope of termination (up to around 24 weeks, though it varies by state) be allowed to terminate her pregnancy for any reason whatsoever? Or must she have a so-called "legitimate" rationale?
"As the technical dilemmas get fixed, some of the ethical dilemmas get fixed. But others arise. It's kind of like ethical whack-a-mole," Snyder said.
One of the newer moles to emerge is, if one can fix a damaged gene, for how long should it be fixed? In one child? In the family's whole line, going forward? If the editing is done in the embryo right after the egg and sperm have united and before the cells begin dividing and becoming specialized, when, say, there are just two or four cells, it will likely affect that child's entire reproductive system and thus all of that child's progeny going forward.
"This notion of changing things forever is a major debate," Snyder said. "It literally gets into metaphysics. On the one hand, you could say, well, wouldn't it be great to get rid of Cystic fibrosis forever? What bad could come of getting rid of a mutant gene forever? But we're not smart enough to know what other things the gene might be doing, and how disrupting one thing could affect this network."
As with any tool, there are risks and benefits, said Michael Kalichman, Director of the Research Ethics Program at the University of California San Diego. While we can envision diverse benefits from a better understanding of human biology and medicine, it is clear that our species can also misuse those tools – from stigmatizing children with certain genetic traits as being "less than," aka dystopian sci-fi movies like Gattaca, to judging parents for making sure their child carries or doesn't carry a particular trait.
"The best chance to ensure that the benefits of this technology will outweigh the risks," Kalichman said, "is for all stakeholders to engage in thoughtful conversations, strive for understanding of diverse viewpoints, and then develop strategies and policies to protect against those uses that are considered to be problematic."
Scientists are making machines, wearable and implantable, to act as kidneys
Recent advancements in engineering mean that the first preclinical trials for an artificial kidney could happen soon.
Like all those whose kidneys have failed, Scott Burton’s life revolves around dialysis. For nearly two decades, Burton has been hooked up (or, since 2020, has hooked himself up at home) to a dialysis machine that performs the job his kidneys normally would. The process is arduous, time-consuming, and expensive. Except for a brief window before his body rejected a kidney transplant, Burton has depended on machines to take the place of his kidneys since he was 12-years-old. His whole life, the 39-year-old says, revolves around dialysis.
“Whenever I try to plan anything, I also have to plan my dialysis,” says Burton says, who works as a freelance videographer and editor. “It’s a full-time job in itself.”
Many of those on dialysis are in line for a kidney transplant that would allow them to trade thrice-weekly dialysis and strict dietary limits for a lifetime of immunosuppressants. Burton’s previous transplant means that his body will likely reject another donated kidney unless it matches perfectly—something he’s not counting on. It’s why he’s enthusiastic about the development of artificial kidneys, small wearable or implantable devices that would do the job of a healthy kidney while giving users like Burton more flexibility for traveling, working, and more.
Still, the devices aren’t ready for testing in humans—yet. But recent advancements in engineering mean that the first preclinical trials for an artificial kidney could happen soon, according to Jonathan Himmelfarb, a nephrologist at the University of Washington.
“It would liberate people with kidney failure,” Himmelfarb says.
An engineering marvel
Compared to the heart or the brain, the kidney doesn’t get as much respect from the medical profession, but its job is far more complex. “It does hundreds of different things,” says UCLA’s Ira Kurtz.
Kurtz would know. He’s worked as a nephrologist for 37 years, devoting his career to helping those with kidney disease. While his colleagues in cardiology and endocrinology have seen major advances in the development of artificial hearts and insulin pumps, little has changed for patients on hemodialysis. The machines remain bulky and require large volumes of a liquid called dialysate to remove toxins from a patient’s blood, along with gallons of purified water. A kidney transplant is the next best thing to someone’s own, functioning organ, but with over 600,000 Americans on dialysis and only about 100,000 kidney transplants each year, most of those in kidney failure are stuck on dialysis.
Part of the lack of progress in artificial kidney design is the sheer complexity of the kidney’s job. Each of the 45 different cell types in the kidney do something different.
Part of the lack of progress in artificial kidney design is the sheer complexity of the kidney’s job. To build an artificial heart, Kurtz says, you basically need to engineer a pump. An artificial pancreas needs to balance blood sugar levels with insulin secretion. While neither of these tasks is simple, they are fairly straightforward. The kidney, on the other hand, does more than get rid of waste products like urea and other toxins. Each of the 45 different cell types in the kidney do something different, helping to regulate electrolytes like sodium, potassium, and phosphorous; maintaining blood pressure and water balance; guiding the body’s hormonal and inflammatory responses; and aiding in the formation of red blood cells.

There's been little progress for patients during Ira Kurtz's 37 years as a nephrologist. Artificial kidneys would change that.
UCLA
Dialysis primarily filters waste, and does so well enough to keep someone alive, but it isn’t a true artificial kidney because it doesn’t perform the kidney’s other jobs, according to Kurtz, such as sensing levels of toxins, wastes, and electrolytes in the blood. Due to the size and water requirements of existing dialysis machines, the equipment isn’t portable. Physicians write a prescription for a certain duration of dialysis and assess how well it’s working with semi-regular blood tests. The process of dialysis itself, however, is conducted blind. Doctors can’t tell how much dialysis a patient needs based on kidney values at the time of treatment, says Meera Harhay, a nephrologist at Drexel University in Philadelphia.
But it’s the impact of dialysis on their day-to-day lives that creates the most problems for patients. Only one-quarter of those on dialysis are able to remain employed (compared to 85% of similar-aged adults), and many report a low quality of life. Having more flexibility in life would make a major different to her patients, Harhay says.
“Almost half their week is taken up by the burden of their treatment. It really eats away at their freedom and their ability to do things that add value to their life,” she says.
Art imitates life
The challenge for artificial kidney designers was how to compress the kidney’s natural functions into a portable, wearable, or implantable device that wouldn’t need constant access to gallons of purified and sterilized water. The other universal challenge they faced was ensuring that any part of the artificial kidney that would come in contact with blood was kept germ-free to prevent infection.
As part of the 2021 KidneyX Prize, a partnership between the U.S. Department of Health and Human Services and the American Society of Nephrology, inventors were challenged to create prototypes for artificial kidneys. Himmelfarb’s team at the University of Washington’s Center for Dialysis Innovation won the prize by focusing on miniaturizing existing technologies to create a portable dialysis machine. The backpack sized AKTIV device (Ambulatory Kidney to Increase Vitality) will recycle dialysate in a closed loop system that removes urea from blood and uses light-based chemical reactions to convert the urea to nitrogen and carbon dioxide, which allows the dialysate to be recirculated.
Himmelfarb says that the AKTIV can be used when at home, work, or traveling, which will give users more flexibility and freedom. “If you had a 30-pound device that you could put in the overhead bins when traveling, you could go visit your grandkids,” he says.
Kurtz’s team at UCLA partnered with the U.S. Kidney Research Corporation and Arkansas University to develop a dialysate-free desktop device (about the size of a small printer) as the first phase of a progression that will he hopes will lead to something small and implantable. Part of the reason for the artificial kidney’s size, Kurtz says, is the number of functions his team are cramming into it. Not only will it filter urea from blood, but it will also use electricity to help regulate electrolyte levels in a process called electrodeionization. Kurtz emphasizes that these additional functions are what makes his design a true artificial kidney instead of just a small dialysis machine.
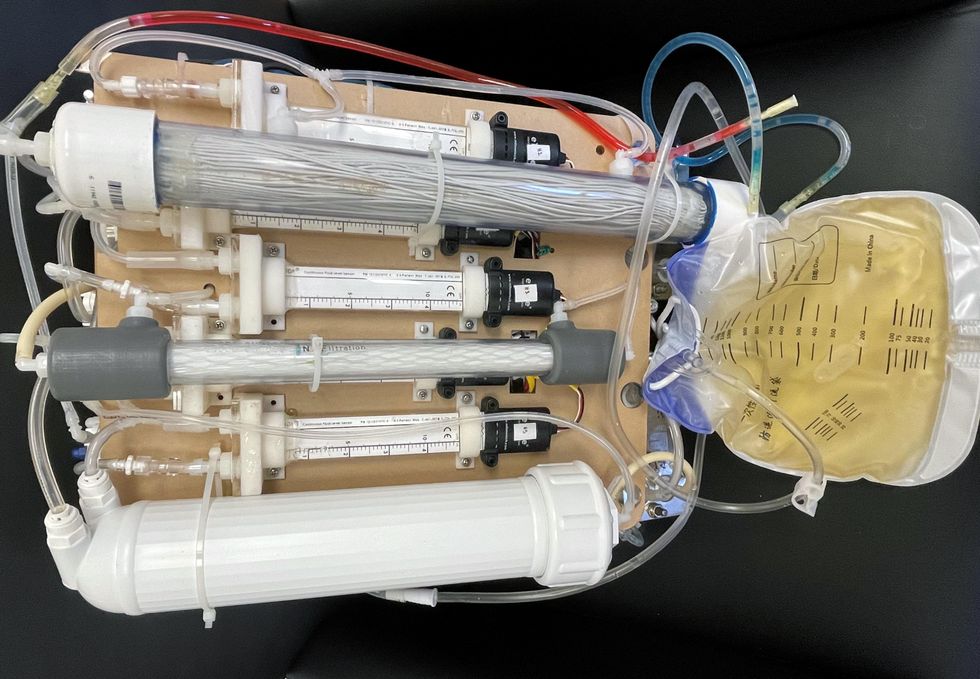
One version of an artificial kidney.
UCLA
“It doesn't have just a static function. It has a bank of sensors that measure chemicals in the blood and feeds that information back to the device,” Kurtz says.
Other startups are getting in on the game. Nephria Bio, a spinout from the South Korean-based EOFlow, is working to develop a wearable dialysis device, akin to an insulin pump, that uses miniature cartridges with nanomaterial filters to clean blood (Harhay is a scientific advisor to Nephria). Ian Welsford, Nephria’s co-founder and CTO, says that the device’s design means that it can also be used to treat acute kidney injuries in resource-limited settings. These potentials have garnered interest and investment in artificial kidneys from the U.S. Department of Defense.
For his part, Burton is most interested in an implantable device, as that would give him the most freedom. Even having a regular outpatient procedure to change batteries or filters would be a minor inconvenience to him.
“Being plugged into a machine, that’s not mimicking life,” he says.
This article was first published by Leaps.org on May 5, 2022.
With this new technology, hospitals and pharmacies could make vaccines and medicines onsite
New research focuses on methods that could change medicine-making worldwide. The scientists propose bursting cells open, removing their DNA and using the cellular gears inside to make therapies.
Most modern biopharmaceutical medicines are produced by workhorse cells—typically bacterial but sometimes mammalian. The cells receive the synthesizing instructions on a snippet of a genetic code, which they incorporate into their DNA. The cellular machinery—ribosomes, RNAs, polymerases, and other compounds—read and use these instructions to build the medicinal molecules, which are harvested and administered to patients.
Although a staple of modern pharma, this process is complex and expensive. One must first insert the DNA instructions into the cells, which they may or may not uptake. One then must grow the cells, keeping them alive and well, so that they produce the required therapeutics, which then must be isolated and purified. To make this at scale requires massive bioreactors and big factories from where the drugs are distributed—and may take a while to arrive where they’re needed. “The pandemic showed us that this method is slow and cumbersome,” says Govind Rao, professor of biochemical engineering who directs the Center for Advanced Sensor Technology at the University of Maryland, Baltimore County (UMBC). “We need better methods that can work faster and can work locally where an outbreak is happening.”
Rao and his team of collaborators, which spans multiple research institutions, believe they have a better approach that may change medicine-making worldwide. They suggest forgoing the concept of using living cells as medicine-producers. Instead, they propose breaking the cells and using the remaining cellular gears for assembling the therapeutic compounds. Instead of inserting the DNA into living cells, the team burst them open, and removed their DNA altogether. Yet, the residual molecular machinery of ribosomes, polymerases and other cogwheels still functioned the way it would in a cell. “Now if you drop your DNA drug-making instructions into that soup, this machinery starts making what you need,” Rao explains. “And because you're no longer worrying about living cells, it becomes much simpler and more efficient.” The collaborators detail their cell-free protein synthesis or CFPS method in their recent paper published in preprint BioAxiv.
While CFPS does not use living cells, it still needs the basic building blocks to assemble proteins from—such as amino acids, nucleotides and certain types of enzymes. These are regularly added into this “soup” to keep the molecular factory chugging. “We just mix everything in as a batch and we let it integrate,” says James Robert Swartz, professor of chemical engineering and bioengineering at Stanford University and co-author of the paper. “And we make sure that we provide enough oxygen.” Rao likens the process to making milk from milk powder.
For a variety of reasons—from the field’s general inertia to regulatory approval hurdles—the method hasn’t become mainstream. The pandemic rekindled interest in medicines that can be made quickly and easily, so it drew more attention to the technology.
The idea of a cell-free protein synthesis is older than one might think. Swartz first experimented with it around 1997, when he was a chemical engineer at Genentech. While working on engineering bacteria to make pharmaceuticals, he discovered that there was a limit to what E. coli cells, the workhorse darling of pharma, could do. For example, it couldn’t grow and properly fold some complex proteins. “We tried many genetic engineering approaches, many fermentation, development, and environmental control approaches,” Swartz recalls—to no avail.
“The organism had its own agenda,” he quips. “And because everything was happening within the organism, we just couldn't really change those conditions very easily. Some of them we couldn’t change at all—we didn’t have control.”
It was out of frustration with the defiant bacteria that a new idea took hold. Could the cells be opened instead, so that the protein-forming reactions could be influenced more easily? “Obviously, we’d lose the ability for them to reproduce,” Swartz says. But that also meant that they no longer needed to keep the cells alive and could focus on making the specific reactions happen. “We could take the catalysts, the enzymes, and the more complex catalysts and activate them, make them work together, much as they would in a living cell, but the way we wanted.”
In 1998, Swartz joined Stanford, and began perfecting the biochemistry of the cell-free method, identifying the reactions he wanted to foster and stopping those he didn’t want. He managed to make the idea work, but for a variety of reasons—from the field’s general inertia to regulatory approval hurdles—the method hasn’t become mainstream. The pandemic rekindled interest in medicines that can be made quickly and easily, so it drew more attention to the technology. For their BioArxiv paper, the team tested the method by growing a specific antiviral protein called griffithsin.
First identified by Barry O’Keefe at National Cancer Institute over a decade ago, griffithsin is an antiviral known to interfere with many viruses’ ability to enter cells—including HIV, SARS, SARS-CoV-2, MERS and others. Originally isolated from the red algae Griffithsia, it works differently from antibodies and antibody cocktails.
Most antiviral medicines tend to target the specific receptors that viruses use to gain entry to the cells they infect. For example, SARS-CoV-2 uses the infamous spike protein to latch onto the ACE2 receptor of mammalian cells. The antibodies or other antiviral molecules stick to the spike protein, shutting off its ability to cling onto the ACE2 receptors. Unfortunately, the spike proteins mutate very often, so the medicines lose their potency. On the contrary, griffithsin has the ability to cling to the different parts of viral shells called capsids—namely to the molecules of mannose, a type of sugar. That extra stuff, glued all around the capsid like dead weight, makes it impossible for the virus to squeeze into the cell.
“Every time we have a vaccine or an antibody against a specific SARS-CoV-2 strain, that strain then mutates and so you lose efficacy,” Rao explains. “But griffithsin molecules glom onto the viral capsid, so the capsid essentially becomes a sticky mess and can’t enter the cell.” Mannose molecules also don’t mutate as easily as viruses’ receptors, so griffithsin-based antivirals do not have to be constantly updated. And because mannose molecules are found on many viruses’ capsids, it makes griffithsin “a universal neutralizer,” Rao explains.
“When griffithsin was discovered, we recognized that it held a lot of promise as a potential antiviral agent,” O’Keefe says. In 2010, he published a paper about griffithsin efficacy in neutralizing viruses of the corona family—after the first SARS outbreak in the early 2000s, the scientific community was interested in such antivirals. Yet, griffithsin is still not available as an off-the-shelf product. So during the Covid pandemic, the team experimented with synthesizing griffithsin using the cell-free production method. They were able to generate potent griffithsin in less than 24 hours without having to grow living cells.
The antiviral protein isn't the only type of medicine that can be made cell-free. The proteins needed for vaccine production could also be made the same way. “Such portable, on-demand drug manufacturing platforms can produce antiviral proteins within hours, making them ideal for combating future pandemics,” Rao says. “We would be able to stop the pandemic before it spreads.”
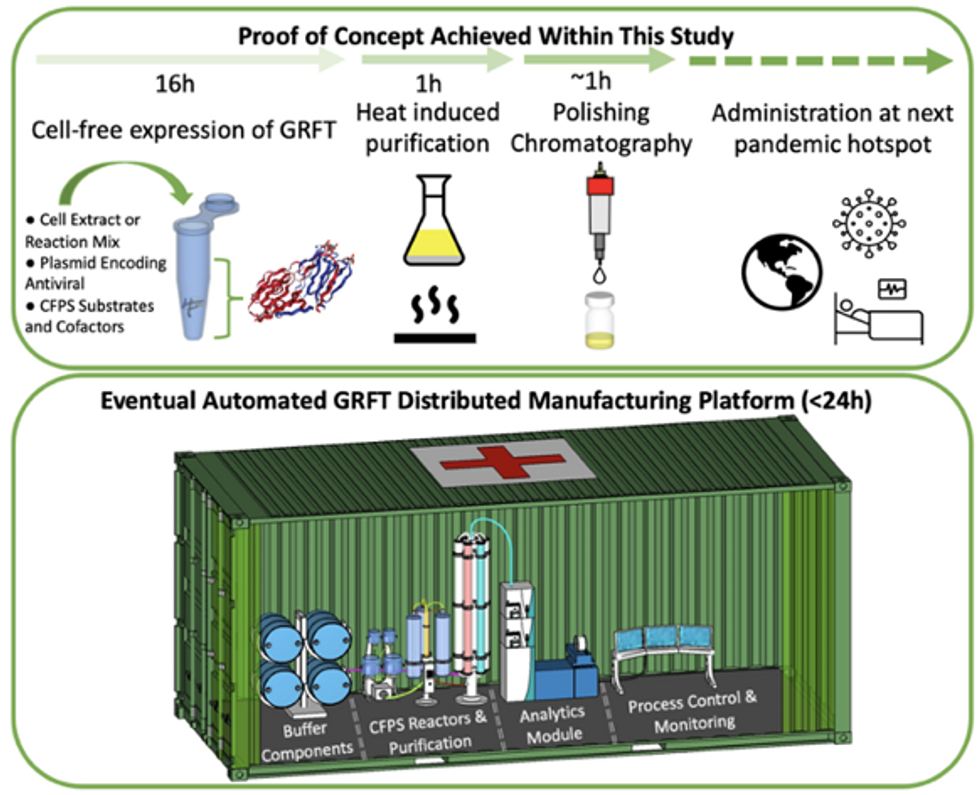
Top: Describes the process used in the study. Bottom: Describes how the new medicines and vaccines could be made at the site of a future viral outbreak.
Image courtesy of Rao and team, sourced from An approach to rapid distributed manufacturing of broad spectrumanti-viral griffithsin using cell-free systems to mitigate pandemics.
Rao’s idea is to perfect the technology to the point that any hospital or pharmacy can load up the media containing molecular factories, mix up the required amino acids, nucleotides and enzymes, and harvest the meds within hours. That will allow making medicines onsite and on demand. “That would be a self-contained production unit, so that you could just ship the production wherever the pandemic is breaking out,” says Swartz.
These units and the meds they produce, will, of course, have to undergo rigorous testing. “The biggest hurdles will be validating these against conventional technology,” Rao says. The biotech industry is risk-averse and prefers the familiar methods. But if this approach works, it may go beyond emergency situations and revolutionize the medicine-making paradigm even outside hospitals and pharmacies. Rao hopes that someday the method might become so mainstream that people may be able to buy and operate such reactors at home. “You can imagine a diabetic patient making insulin that way, or some other drugs,” Rao says. It would work not unlike making baby formula from the mere white powder. Just add water—and some oxygen, too.
Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.