New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
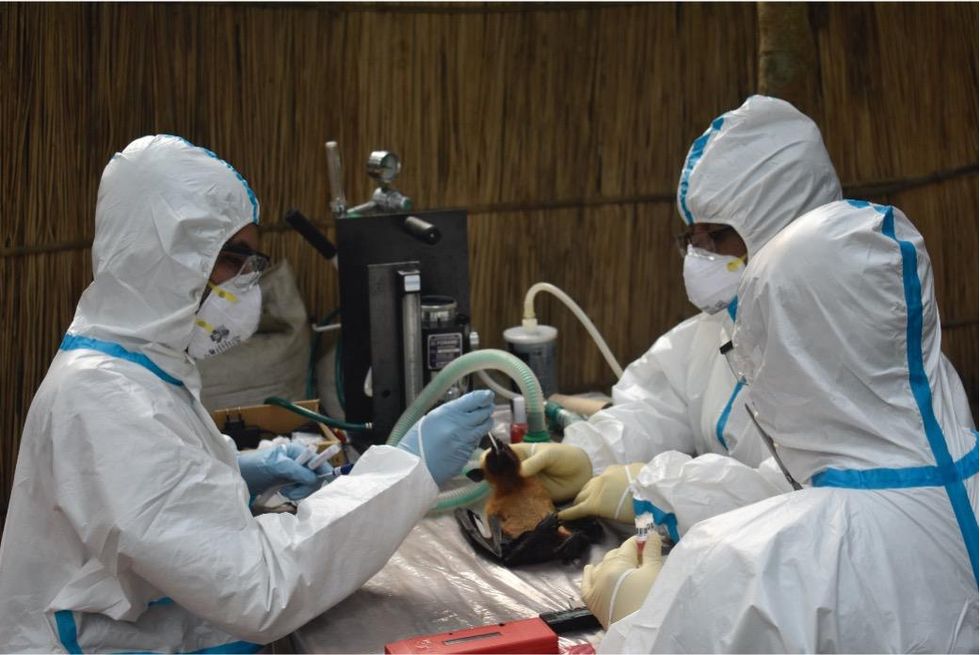
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
This man spent over 70 years in an iron lung. What he was able to accomplish is amazing.
Paul Alexander spent more than 70 years confined to an iron lung after a polio infection left him paralyzed at age 6. Here, Alexander uses a mirror attached to the top of his iron lung to view his surroundings.
It’s a sight we don’t normally see these days: A man lying prone in a big, metal tube with his head sticking out of one end. But it wasn’t so long ago that this sight was unfortunately much more common.
In the first half of the 20th century, tens of thousands of people each year were infected by polio—a highly contagious virus that attacks nerves in the spinal cord and brainstem. Many people survived polio, but a small percentage of people who did were left permanently paralyzed from the virus, requiring support to help them breathe. This support, known as an “iron lung,” manually pulled oxygen in and out of a person’s lungs by changing the pressure inside the machine.
Paul Alexander was one of several thousand who were infected and paralyzed by polio in 1952. That year, a polio epidemic swept the United States, forcing businesses to close and polio wards in hospitals all over the country to fill up with sick children. When Paul caught polio in the summer of 1952, doctors urged his parents to let him rest and recover at home, since the hospital in his home suburb of Dallas, Texas was already overrun with polio patients.
Paul rested in bed for a few days with aching limbs and a fever. But his condition quickly got worse. Within a week, Paul could no longer speak or swallow, and his parents rushed him to the local hospital where the doctors performed an emergency procedure to help him breathe. Paul woke from the surgery three days later, and found himself unable to move and lying inside an iron lung in the polio ward, surrounded by rows of other paralyzed children.

But against all odds, Paul lived. And with help from a physical therapist, Paul was able to thrive—sometimes for small periods outside the iron lung.
The way Paul did this was to practice glossopharyngeal breathing (or as Paul called it, “frog breathing”), where he would trap air in his mouth and force it down his throat and into his lungs by flattening his tongue. This breathing technique, taught to him by his physical therapist, would allow Paul to leave the iron lung for increasing periods of time.
With help from his iron lung (and for small periods of time without it), Paul managed to live a full, happy, and sometimes record-breaking life. At 21, Paul became the first person in Dallas, Texas to graduate high school without attending class in person, owing his success to memorization rather than taking notes. After high school, Paul received a scholarship to Southern Methodist University and pursued his dream of becoming a trial lawyer and successfully represented clients in court.

Throughout his long life, Paul was also able to fly on a plane, visit the beach, adopt a dog, fall in love, and write a memoir using a plastic stick to tap out a draft on a keyboard. In recent years, Paul joined TikTok and became a viral sensation with more than 330,000 followers. In one of his first videos, Paul advocated for vaccination and warned against another polio epidemic.
Paul was reportedly hospitalized with COVID-19 at the end of February and died on March 11th, 2024. He currently holds the Guiness World Record for longest survival inside an iron lung—71 years.
Polio thankfully no longer circulates in the United States, or in most of the world, thanks to vaccines. But Paul continues to serve as a reminder of the importance of vaccination—and the power of the human spirit.
““I’ve got some big dreams. I’m not going to accept from anybody their limitations,” he said in a 2022 interview with CNN. “My life is incredible.”
When doctors couldn’t stop her daughter’s seizures, this mom earned a PhD and found a treatment herself.
Savannah Salazar (left) and her mother, Tracy Dixon-Salazaar, who earned a PhD in neurobiology in the quest for a treatment of her daughter's seizure disorder.
Twenty-eight years ago, Tracy Dixon-Salazaar woke to the sound of her daughter, two-year-old Savannah, in the midst of a medical emergency.
“I entered [Savannah’s room] to see her tiny little body jerking about violently in her bed,” Tracy said in an interview. “I thought she was choking.” When she and her husband frantically called 911, the paramedic told them it was likely that Savannah had had a seizure—a term neither Tracy nor her husband had ever heard before.
Over the next several years, Savannah’s seizures continued and worsened. By age five Savannah was having seizures dozens of times each day, and her parents noticed significant developmental delays. Savannah was unable to use the restroom and functioned more like a toddler than a five-year-old.
Doctors were mystified: Tracy and her husband had no family history of seizures, and there was no event—such as an injury or infection—that could have caused them. Doctors were also confused as to why Savannah’s seizures were happening so frequently despite trying different seizure medications.
Doctors eventually diagnosed Savannah with Lennox-Gaustaut Syndrome, or LGS, an epilepsy disorder with no cure and a poor prognosis. People with LGS are often resistant to several kinds of anti-seizure medications, and often suffer from developmental delays and behavioral problems. People with LGS also have a higher chance of injury as well as a higher chance of sudden unexpected death (SUDEP) due to the frequent seizures. In about 70 percent of cases, LGS has an identifiable cause such as a brain injury or genetic syndrome. In about 30 percent of cases, however, the cause is unknown.
Watching her daughter struggle through repeated seizures was devastating to Tracy and the rest of the family.
“This disease, it comes into your life. It’s uninvited. It’s unannounced and it takes over every aspect of your daily life,” said Tracy in an interview with Today.com. “Plus it’s attacking the thing that is most precious to you—your kid.”
Desperate to find some answers, Tracy began combing the medical literature for information about epilepsy and LGS. She enrolled in college courses to better understand the papers she was reading.
“Ironically, I thought I needed to go to college to take English classes to understand these papers—but soon learned it wasn’t English classes I needed, It was science,” Tracy said. When she took her first college science course, Tracy says, she “fell in love with the subject.”
Tracy was now a caregiver to Savannah, who continued to have hundreds of seizures a month, as well as a full-time student, studying late into the night and while her kids were at school, using classwork as “an outlet for the pain.”
“I couldn’t help my daughter,” Tracy said. “Studying was something I could do.”
Twelve years later, Tracy had earned a PhD in neurobiology.
After her post-doctoral training, Tracy started working at a lab that explored the genetics of epilepsy. Savannah’s doctors hadn’t found a genetic cause for her seizures, so Tracy decided to sequence her genome again to check for other abnormalities—and what she found was life-changing.
Tracy discovered that Savannah had a calcium channel mutation, meaning that too much calcium was passing through Savannah’s neural pathways, leading to seizures. The information made sense to Tracy: Anti-seizure medications often leech calcium from a person’s bones. When doctors had prescribed Savannah calcium supplements in the past to counteract these effects, her seizures had gotten worse every time she took the medication. Tracy took her discovery to Savannah’s doctor, who agreed to prescribe her a calcium blocker.
The change in Savannah was almost immediate.
Within two weeks, Savannah’s seizures had decreased by 95 percent. Once on a daily seven-drug regimen, she was soon weaned to just four, and then three. Amazingly, Tracy started to notice changes in Savannah’s personality and development, too.
“She just exploded in her personality and her talking and her walking and her potty training and oh my gosh she is just so sassy,” Tracy said in an interview.
Since starting the calcium blocker eleven years ago, Savannah has continued to make enormous strides. Though still unable to read or write, Savannah enjoys puzzles and social media. She’s “obsessed” with boys, says Tracy. And while Tracy suspects she’ll never be able to live independently, she and her daughter can now share more “normal” moments—something she never anticipated at the start of Savannah’s journey with LGS. While preparing for an event, Savannah helped Tracy get ready.
“We picked out a dress and it was the first time in our lives that we did something normal as a mother and a daughter,” she said. “It was pretty cool.”

