New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
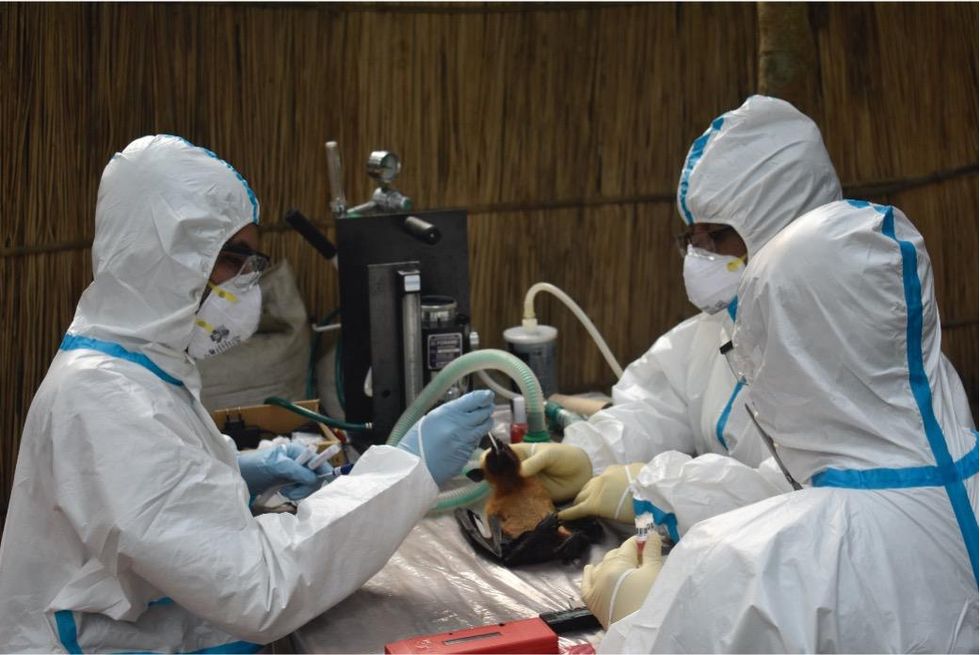
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
The Women of RNA: Two Award-Winners Share Why They Spent Their Careers Studying DNA's Lesser-Known Cousin
When Lynne Maquat, who leads the Center for RNA Biology at the University of Rochester, became interested in the ribonucleic acid molecule in the 1970s, she was definitely in the minority. The same was true for Joan Steitz, now professor of molecular biophysics and biochemistry at Yale University, who began to study RNA a decade earlier in the 1960s.
"My first RNA experiment was a failure, because we didn't understand how things worked," Steitz recalls. In her first undergraduate experiment, she unwittingly used a lab preparation that destroyed the RNA. "Unknowingly, our preparation contained enzymes that degraded our RNA."
At the time, scientists pursuing genetic research tended to focus on DNA, or deoxyribonucleic acid — and for good reason. It was clear that the enigmatic double-helix ribbon held the answers to organisms' heredity, genetic traits, development, growth and aging. If scientists could decipher the secrets of DNA and understand how its genetic instructions translate into the body's functions in health and disease, they could develop treatments for all kinds of diseases. On the contrary, the prevailing dogma of the time viewed RNA as merely a helper that passively carried out DNA's genetic instructions for protein-making — so it received much less attention.
But Maquat and Steitz weren't interested in heredity. They studied biochemistry and biophysics, so they wanted to understand how RNA functioned on the molecular level — how it carried instructions, catalyzed reactions, and helped build protein bonds, among other things.
"I'm a mechanistic biochemist, so I like to know how things happen," Maquat says. "Once you understand the mechanism, you can think of how to solve problems." And so the quest to understand how RNA does its job became the focus of both women's careers.
"People can now appreciate why some of us studied RNA for such a long time."
Half a century later, in 2021, their RNA work has earned two prestigious recognitions only months from each other. In February, they received the Wolf Prize in Medicine, followed by the Warren Alpert Foundation Prize in May, awarded to scientists whose achievements led to prevention, cure or treatments of human diseases.
It was the development of the COVID-19 vaccines that made RNA a household name. Made by Moderna and Pfizer, the vaccines use the RNA molecule to deliver genetic instructions for making SARS-CoV-2's characteristic spike protein in our cells. The presence of this foreign-looking protein triggers the immune system to attack and remember the pathogen. As the vaccines reached the finish line, RNA took center stage, and it was Maquat's and Steitz's research that helped reveal how these molecular cogwheels drive many biological functions within cells.
If you think of a cell as a kingdom, the DNA plays the role of a queen. Like a monarch in a palace, DNA nestles inside the cell's nucleus issuing instructions needed for the cell to function. But no queen can successfully govern without her court, her messengers, and her soldiers, as well as other players that make her kingdom work. That's what RNAs do — they act as the DNA's vassals. They carry instructions for protein assembly, catalyze reactions and supervise many other processes to make sure the cellular kingdom performs as it should.
There are a myriad of these RNA vassals in our cells, and each type has its own specific task. There are messenger RNAs that deliver genetic instructions for protein synthesis from DNA to ribosomes, the cells' protein-making factories. There are ribosomal RNAs that help stitch together amino acids to make proteins. There are transfer RNAs that can bring amino acids to this protein synthesis machine, keeping it going. Then there are circular RNAs that act as sponges, absorbing proteins to help regulate the activity of genes. And that's only the tip of the iceberg when it comes to RNA diversity, researchers say.
"We know what the most abundant and important RNAs are doing," says Steitz. "But there are thousands of different ones, and we still don't have a full knowledge of them."
Critical to RNA's proper functioning is a process called splicing, in which a precursor mRNA is transformed into mature, fully-functional mRNA — a phenomenon that Steitz's work helped elucidate. The splicing process, which takes place in cellular assembly lines, involves removing extra RNA sequences and stringing the remaining RNA pieces together. Steitz found that tiny RNA particles called snRNPs are crucial to this process. They act as handy helpers, finding and removing errant genetic material from the mRNA molecules.
A dysfunctional RNA assembly line leads to diseases, including many cancers. For instance, Steitz found that people with Lupus — an autoimmune disorder — have antibodies that mistakenly attack the little snRNP helpers. She also discovered that when snRNPs don't do their job properly, they can cause what scientists call mis-splicing, producing defective mRNAs.
Fortunately, cells have a built-in quality-control process that can spot and correct these mistakes, which is what Maquat studied in her work. In 1981, she discovered a molecular quality-control system that spots and destroys such incorrectly assembled mRNA. With the cryptic name "nonsense-mediated mRNA decay" or NMD, this process is vital to the health and wellbeing of a cellular kingdom in humans — because splicing mistakes happen far more often than one would imagine.
"We estimate that about a third of our mRNA are mistakes," Maquat says. "And nonsense-mediated mRNA decay cleans up these mistakes." When this quality-control system malfunctions, defective mRNA forge faulty proteins, which mess up the cellular machinery and cause disease, including various forms of cancer.
Scientists' newfound appreciation of RNA opens door to many novel treatments.
Now that the first RNA-based shots were approved, the same principle can be used for create vaccines for other diseases, the two RNA researchers say. Moreover, the molecule has an even greater potential — it can serve as a therapeutic target for other disorders. For example, Spinraza, a groundbreaking drug approved in 2016 for spinal muscular atrophy, uses small snippets of synthetic genetic material that bind to the RNA, helping fix splicing errors. "People can now appreciate why some of us studied RNA for such a long time," says Maquat.
Steitz is thrilled that the entire field of RNA research is enjoying the limelight. "I'm delighted because the prize is more of a recognition of the field than just our work," she says. "This is a more general acknowledgment of how basic research can have a remarkable impact on human health."
Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.
In 2010, a 67-year-old former executive assistant for a Fortune 500 company was diagnosed with mild cognitive impairment. By 2014, her doctors confirmed she had Alzheimer's disease.
As her disease progressed, she continued to live independently but wasn't able to drive anymore. Today, she can manage most of her everyday tasks, but her two daughters are considering a live-in caregiver. Despite her condition, the woman may represent a beacon of hope for the approximately 44 million people worldwide living with Alzheimer's disease. The now 74-year-old is among a small cadre of Alzheimer's patients who have undergone an experimental ultrasound procedure aimed at slowing cognitive decline.
In November 2020, Elisa Konofagou, a professor of biomedical engineering and director of the Ultrasound and Elasticity Imaging Laboratory at Columbia University, and her team used ultrasound to noninvasively open the woman's blood-brain barrier. This barrier is a highly selective membrane of cells that prevents toxins and pathogens from entering the brain while allowing vital nutrients to pass through. This regulatory function means the blood-brain barrier filters out most drugs, making treating Alzheimer's and other brain diseases a challenge.
Ultrasound uses high-frequency sound waves to produce live images from the inside of the human body. But scientists think it could also be used to boost the effectiveness of Alzheimer's drugs, or potentially even improve brain function in dementia patients without the use of drugs.
The procedure, which involves a portable ultrasound system, is the culmination of 17 years of lab work. As part of a small clinical trial, scientists positioned a sensor transmitting ultrasound waves on top of the woman's head while she sat in a chair. The sensor sends ultrasound pulses throughout the target region. Meanwhile, investigators intravenously infused microbubbles into the woman to boost the effects of the ultrasound. Three days after the procedure, scientists scanned her brain so that they could measure the effects of the treatments. Five months later, they took more images of her brain to see if the effects of the treatment lasted.
Promising Signs
After the first brain scan, Konofagou and her team found that amyloid-beta, the protein that clumps together in the brains of Alzheimer's patients and disrupts cell function, had declined by 14%. At the woman's second scan, amyloid levels were still lower than before the experimental treatment, but only by 10% this time. Konofagou thinks repeat ultrasound treatments given early on in the development of Alzheimer's may have the best chance at keeping amyloid plaques at bay.
This reduction in amyloid appeared to halt the woman's cognitive decline, at least temporarily. Following the ultrasound treatment, the woman took a 30-point test used to measure cognitive impairment in Alzheimer's. Her score — 22, indicating mild cognitive impairment — remained the same as before the intervention. Konofagou says this was actually a good sign.
"Typically, every six months an Alzheimer's patient scores two to three points lower, so this is highly encouraging," she says.
Konofagou speculates that the results might have been even more impressive had they applied the ultrasound on a larger section of the brain at a higher frequency. The selected site was just 4 cubic centimeters. Current safety protocols set by the U.S. Food and Drug Administration stipulate that investigators conducting such trials only treat one brain region with the lowest pressure possible.
The Columbia trial is aided by microbubble technology. During the procedure, investigators infused tiny, gas-filled spheres into the woman's veins to enhance the ultrasound reflection of the sound waves.
The big promise of ultrasound is that it could eventually make drugs for Alzheimer's obsolete.
"Ultrasound with microbubbles wakes up immune cells that go on to discard amyloid-beta," Konofagou says. "In this way, we can recover the function of brain neurons, which are destroyed by Alzheimer's in a sort of domino effect." What's more, a drug delivered alongside ultrasound can penetrate the brain at a dose up to 10 times higher.
Costas Arvanitis, an assistant professor at Georgia Institute of Technology who studies ultrasonic biophysics and isn't involved in the Columbia trial, is excited about the research. "First, by applying ultrasound you can make larger drugs — picture an antibody — available to the brain," he says. Then, you can use ultrasound to improve the therapeutic index, or the ratio of the effectiveness of a drug versus the ratio of adverse effects. "Some drugs might be effective but because we have to provide them in high doses to see significant responses they tend to come with side effects. By improving locally the concentration of a drug, you open up the possibility to reduce the dose."
The Columbia trial will enroll just six patients and is designed to test the feasibility and safety of the approach, not its efficacy. Still, Arvantis is hopeful about the potential benefits of the technique. "The technology has already been demonstrated to be safe, its components are now tuned to the needs of this specific application, and it's safe to say it's only a matter of time before we are able to develop personalized treatments," he says.
Konofagou and her colleagues recently presented their findings at the 20th Annual International Symposium for Therapeutic Ultrasound and intend to publish them in a scientific journal later this year. They plan to recruit more participants for larger trials, which will determine how effective the therapy is at improving memory and brain function in Alzheimer's patients. They're also in talks with pharmaceutical companies about ways to use their therapeutic approach to improve current drugs or even "create new drugs," says Konofagou.
A New Treatment Approach
On June 7, the FDA approved the first Alzheimer's disease drug in nearly two decades. Aducanumab, a drug developed by Biogen, is an antibody designed to target and reduce amyloid plaques. The drug has already sparked immense enthusiasm — and controversy. Proponents say the drug is a much-needed start in the fight against the disease, but others argue that the drug doesn't substantially improve cognition. They say the approval could open the door to the FDA greenlighting more Alzheimer's drugs that don't have a clear benefit, giving false hope to both patients and their families.
Konofagou's ultrasound approach could potentially boost the effects of drugs like aducanumab. "Our technique can be seamlessly combined with aducanumab in early Alzheimer's, where it has shown the most promise, to further enhance both its amyloid load reduction and further reduce cognitive deficits while using exactly the same drug regimen otherwise," she says. For the Columbia team, the goal is to use ultrasound to maximize the effects of aducanumab, as they've done with other drugs in animal studies.
But Konofagou's approach could transcend drug controversies, and even drugs altogether. The big promise of ultrasound is that it could eventually make drugs for Alzheimer's obsolete.
"There are already indications that the immune system is alerted each time ultrasound is exerted on the brain or when the brain barrier is being penetrated and gets activated, which on its own may have sufficient therapeutic effects," says Konofagou. Her team is now working with psychiatrists in hopes of using brain stimulation to treat patients with depression.
The potential to modulate the brain without drugs is huge and untapped, says Kim Butts Pauly, a professor of radiology, electrical engineering and bioengineering at Stanford University, who's not involved in the Columbia study. But she admits that scientists don't know how to fully control ultrasound in the brain yet. "We're only at the starting point of getting the tools to understand and harness how ultrasound microbubbles stimulate an immune response in the brain."
Meanwhile, the 74-year-old woman who received the ultrasound treatment last year, goes on about her life, having "both good days and bad days," her youngest daughter says. COVID-19's isolation took a toll on her, but both she and her daughters remain grateful for the opportunity to participate in the ultrasound trial.
"My mother wants to help, if not for herself, then for those who will follow her," the daughter says. She hopes her mother will be able to join the next phase of the trial, which will involve a drug in conjunction with the ultrasound treatment. "This may be the combination where the magic will happen," her daughter says.

