New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
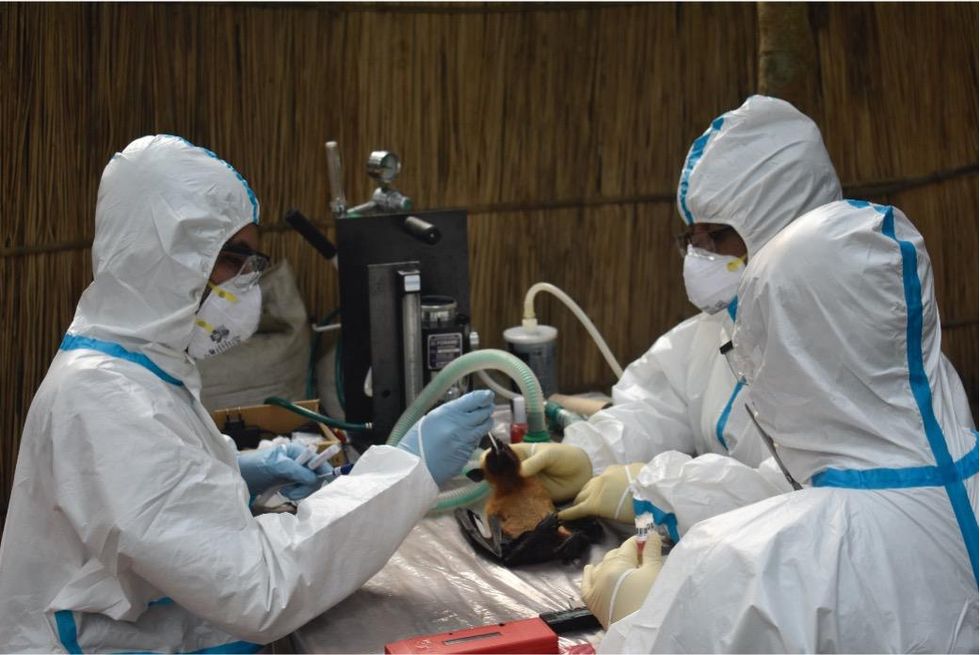
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
A Doctor Who Treated His Own Rare Disease Is Tracking COVID-19 Treatments Hiding In Plain Sight
Dr. David Fajgenbaum looking through a microscope at his lab.
In late March, just as the COVID-19 pandemic was ramping up in the United States, David Fajgenbaum, a physician-scientist at the University of Pennsylvania, devised a 10-day challenge for his lab: they would sift through 1,000 recently published scientific papers documenting cases of the deadly virus from around the world, pluck out the names of any drugs used in an attempt to cure patients, and track the treatments and their outcomes in a database.
Before late 2019, no one had ever had to treat this exact disease before, which meant all treatments would be trial and error. Fajgenbaum, a pioneering researcher in the field of drug repurposing—which prioritizes finding novel uses for existing drugs, rather than arduously and expensively developing new ones for each new disease—knew that physicians around the world would be embarking on an experimental journey, the scale of which would be unprecedented. His intention was to briefly document the early days of this potentially illuminating free-for-all, as a sidebar to his primary field of research on a group of lymph node disorders called Castleman disease. But now, 11 months and 29,000 scientific papers later, he and his team of 22 are still going strong.
They're running a publicly accessible database called the CORONA Project (COvid19 Registry of Off-label & New Agents) that to date tracks 400 different COVID-19 treatments that have been tried somewhere in the world, along with the frequency of their use, and the outcomes.
"There's so many drugs being used all over the place, in different ways, with different outcomes," says Fajgenbaum. "We're trying to add some order to the madness."
20,000 people have accessed the registry—other physicians and researchers, those in the pharmaceutical industry, and even curious lay people—and the data are now being shared with the U.S. Food and Drug Administration in the hopes of launching large-scale trials that would lead to approving a constellation of treatment options for COVID-19 faster than any new drugs could come online.
"What David's group has done with the CORONA Project is on a scale that I don't think has ever been seen before," says Heather Stone, a health science policy analyst at the FDA who specializes in drug repurposing. She was not involved in establishing the project, but is now working with its data. "To collect and collate that information and make it openly accessible is a massive feat, and a huge benefit to the medical community," she says.
On a Personal Mission
In the science and medical world, Fajgenbaum lives a dual existence: he is both researcher and subject, physician and patient. In July 2010, when he was a healthy and physically fit 25-year-old finishing medical school, he began living through what would become a recurring, unprovoked, and overzealous immune response that repeatedly almost killed him.
His lymph nodes were inflamed; his liver, kidneys, and bone marrow were faltering; and he was dead tired all the time. At first his doctors mistook his mysterious illness for lymphoma, but his inflamed lymph nodes were merely a red herring. A month after his initial hospitalization, pathologists at Mayo Clinic finally diagnosed him with idiopathic multicentric Castleman disease—a particularly ruthless form of a class of lymph node disorders that doesn't just attack one part of the body, but many, and has no known cause. It's a rare diagnosis within an already rare set of disorders. Only about 1,500 Americans a year receive the same diagnosis.
Without many options for treatment, Fajgenbaum underwent recurring rounds of chemotherapy. Each time, the treatment would offer temporary respite from Castleman symptoms, but bring the full spate of chemotherapy side effects. And it wasn't a sustainable treatment for the long haul. Regularly dousing a person's cells in unmitigated toxicity was about as elegant a solution to Fajgenbaum's disease as bulldozing a house in response to a toaster fire. The fire might go out (though not necessarily), but the house would be destroyed.
A swirl of exasperation and doggedness finally propelled Fajgenbaum to take on a crucial question himself: Among all of the already FDA-approved drugs on the market, was there something out there, labeled for another use, that could beat back Castleman disease and that he could tolerate long-term? After months of research, he discovered the answer: sirolimus, a drug normally prescribed to patients receiving a kidney transplant, could be used to suppress his overactive immune system with few known side effects to boot.
Fajgenbaum became hellbent on devoting his practice and research to making similar breakthroughs for others. He founded the Castleman Disease Collaborative Network, to coordinate the research of others studying this bewildering disease, and directs a laboratory consumed with studying cytokine storms—out-of-control immune responses characterized by the body's release of cytokines, proteins that the immune system secretes and uses to communicate with and direct other cells.
In the spring of 2020, when cytokine storms emerged as a hallmark of the most severe and deadly cases of COVID-19, Fajgenbaum's ears perked up. Although SARS-CoV-2 itself was novel, Fajgenbaum already had almost a decade of experience battling the most severe biological forces it brought. Only this time, he thought, it might actually be easier to pinpoint a treatment—unlike Castleman disease, which has no known cause, at least here a virus was clearly the instigator.
"Because [a drug] looks promising, we need to do a well-designed, large randomized controlled trial to really investigate whether this drug works or not ... We don't use that to say, 'You should take it.'"
Thinking Beyond COVID
The week of March 13, when the World Health Organization declared COVID-19 a pandemic, Fajgenbaum found himself hoping that someone would make the same connection and apply the research to COVID. "Then like a minute later I was like, 'Why am I hoping that someone, somewhere, either follows our footsteps, or has a similar background to us? Maybe we just need to do it," he says. And the CORONA Project was born—first as a 10-day exercise, and later as the robust, interactive tool it now is.
All of the 400 treatments in the CORONA database are examples of repurposed drugs, or off-label uses: physicians are prescribing drugs to treat COVID that have been approved for a different disease. There are no bonafide COVID treatments, only inferences. The goal for people like Fajgenbaum and Stone is to identify potential treatments for further study and eventual official approval, so that physicians can treat the disease with a playbook in hand. When it works, drug repurposing opens up a way to move quickly: A range of treatments could be available to patients within just a few years of a totally new virus entering our reality compared with the 12 - 19 years new drug development takes.
"Companies for many decades have explored the use of their products for not just a single indication but often for many indications," says Stone. "'Supplemental approvals' are all essentially examples of drug repurposing, we just didn't call it that. The challenge, I think, is to explore those opportunities more comprehensively and systematically to really try to understand the full breadth of potential activity of any drug or molecule."
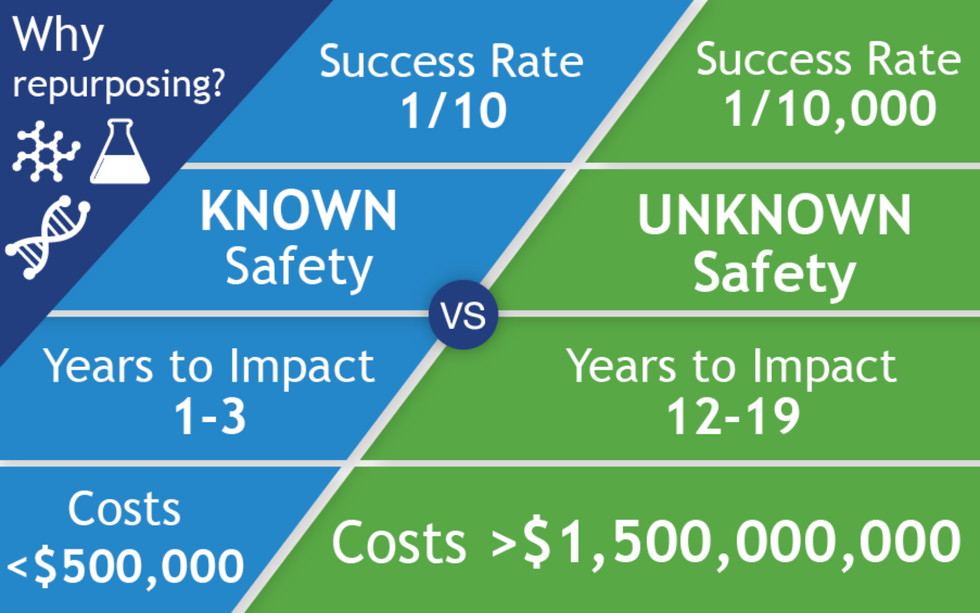
The left column shows the path of a repurposed drug, and on the right is the path of a newly discovered and developed drug.
Cures Within Reach
In Fajgenbaum's primary work, promising drugs stand out easily. For a disease like Castleman, where improvement almost never occurs on its own, any improvement that follows a treatment can pretty clearly be attributed to that treatment. But Fajgenbaum says tracking COVID outcomes is less straightforward since "the vast majority of people will get better, whether they take steroids or they take Skittles." That's why the intent of the database is to identify promising treatments only to generate hypotheses and fruitful clinical trials, not to offer full-throated treatment recommendations. Within the registry, Fajgenbaum considers a drug promising if it's being used in humans, not just in lab animals, and a significant proportion of cases report patient improvement.
"It's that sort of combination of rock-solid randomized controlled trial data, plus anecdotal retrospective data, that we combine to say, 'Wow, this drug looks more promising than another,'" says Fajgenbaum. "Because it looks promising, we need to do a well-designed, large randomized controlled trial to really investigate whether this drug works or not ... We don't use that to say, 'You should take it.'"
Experts say that the search for repurposed drugs to treat COVID could have implications for rare diseases in general. Rare diseases, of which Castleman is one, affect 400 million people around the world. 95% of them don't have a tailor-made, FDA-approved drug treatment. Developing one is a lengthy and often prohibitively expensive process. If only a dozen people will benefit from and buy a drug, it's not often worth it to pharmaceutical companies to spend millions of dollars making them. On occasion when they do, however, that overhead shows up in the price tag: the top 10 most expensive drugs in the world are all for rare diseases, often making them inaccessible to patients. Identifying new clinical uses for drugs that already exist is critical for opening a trap door out of a cycle that prioritizes profits over health outcomes.
"COVID is an interesting case where it's demonstrated that when the scientific and medical community really focuses all of its efforts and talents on a single problem, a solution can be identified and in a much faster time period than has ever historically been the case," says Stone. "I certainly wish it hadn't taken a pandemic to do that, but I think it does have lessons for the future in terms of our ability to accomplish things that we might have previously not thought were possible"—for example, mainstreaming the idea of drug repurposing as a treatment tool, even long after the pandemic subsides.
A Confounding Virus
The FDA declined to comment on what drugs it was fast-tracking for trials, but Fajgenbaum says that based on the CORONA Project's data, which includes data from smaller trials that have already taken place, he feels there are three drugs that seem the most clearly and broadly promising for large-scale studies. Among them are dexamethasone, which is a steroid with anti-inflammatory effects, and baricitinib, a rheumatoid arthritis drug, both of which have enabled the sickest COVID-19 patients to bounce back by suppressing their immune systems. The third most clearly promising drug is heparin, a blood thinner, which a recent trial showed to be most helpful when administered at a full dose, more so than at a small, preventative dose. (On the flipside, Fajgenbaum says "it's a little sad" that in the database you can see hydroxychloroquine is still the most-prescribed drug being tried as a COVID treatment around the world, despite over the summer being debunked widely as an effective treatment, and continuously since then.)
One of the confounding attributes of SARS-CoV-2 is its ability to cause such a huge spectrum of outcomes. It's unlikely a silver bullet treatment will emerge under that reality, so the database also helps surface drugs that seem most promising for a specific population. Fluvoxamine, a selective serotonin reuptake inhibitor used to treat obsessive compulsive disorder, showed promise in the recovery of outpatients—those who were sick, but not severely enough to be hospitalized. Tocilizumab, which was actually developed for Castleman disease, the disease Fajgenbaum is managing, was initially written off as a COVID treatment because it failed to benefit large portions of hospitalized patients, but now seems to be effective if used on intensive care unit patients within 24 hours of admission—these are some of the sickest patients with the highest risk of dying.
Other than fluvoxamine, most of the drugs labeled as promising do skew toward targeting hospitalized patients, more than outpatients. One reason, Fajgenbaum says, is that "if you're in a hospital it's very easy to give you a drug and to track you, and there are very objective measurements as to whether you die, you progress to a ventilator, etc." Tracking outpatients is far more difficult, especially when folks have been routinely asked to stay home, quarantine, and free up hospital resources if they're experiencing only mild symptoms.
But the other reason for the skew is because COVID is very unlike most other diseases in terms of the human immune response the virus triggers. For example, if oncology treatments show some benefit to people with the highest risk of dying, then they usually work extremely well if administered in the earlier stages of a cancer diagnosis. Across many diseases, this dialing backward is a standard approach to identifying promising treatments. With COVID, all of that reasoning has proven moot.
As we've seen over the last year, COVID cases often start as asymptomatic, and remain that way for days, indicating the body is mounting an incredibly weak immune response initially. Then, between days five and 14, as if trying to make up for lost time, the immune system overcompensates by launching a major inflammatory response, which in the sickest patient can lead to the type of cytokine storms that helped Fajgenbaum realize his years of Castleman research might be useful during this public health crisis. Because of this phased response, you can't apply the same treatment logic to all cases.
"In COVID, drugs that work late tend to not work if given early, and drugs that work early tend to not work if given late," says Fajgenbaum. "Generally this … is not a commonplace thing for a virus."
"There are drugs that are literally sitting in every single hospital pharmacy in the country that, if a study shows it's effective, can be deployed that evening to patients on a massive scale."
This see-sawing necessitates tracking a constellation of drugs that might work for different stages of the disease as a patient moves from the weak immune response stage into the overzealous immune response.
"COVID is difficult, compared to other diseases, because there are so many different levels of disease severity, and recovery at different rates," says Stone, the FDA researcher. "That makes it hard to see the patterns or signals and it makes it very important to collect very, very large numbers of cases in order to really reliably identify signals."
This particular moment in the pandemic feels like a massive tipping point, or the instant a tiny pinprick of light finally appeared at the end of the tunnel: several vaccines are already here, with more on the way imminently. In the U.S., more than 65 million doses of the vaccine have been administered, and positive COVID cases are finally falling back to levels not seen since October. On the hopeful surface, it might seem a strange moment to be preparing to launch trials that will validate treatments for a virus it seems the U.S. may finally be beating back. But at best, Americans are still months away from reaching herd immunity through vaccination, and new circulating variants may threaten to upend our fragile progress.
"In the meantime, there are drugs that are literally sitting in every single hospital pharmacy in the country that, if a study shows it's effective, can be deployed that evening to patients on a massive scale. It wouldn't have to be newly produced, it wouldn't have to be shipped, it's literally there already," says Fajgenbaum. "The idea that you can save a lot of lives by finding things that are just already there I think is really compelling, given how many people are going to die over these next few months."
Even after that, not everyone can or will be vaccinated, and, as the Wall Street Journal recently reported, "The pathogen will circulate for years, or even decades, leaving society to coexist with Covid-19 much as it does with other endemic diseases like flu, measles, and HIV." Neither vaccines, personal behavior, or treatments alone is a panacea against the virus, but together they might be.
"It's important to explore all avenues in this public health emergency, and drug repurposing can continue to play a role as the pandemic continues and evolves," says Stone. "I think COVID variants in particular are a big concern at the moment, and therefore continuing to investigate new therapeutics, even as the vaccines roll out, will continue to be a priority."
How a Nobel-Prize Winner Fought Her Family, Nazis, and Bombs to Change our Understanding of Cells Forever
Rita Levi-Montalcini survived the Nazis and eventually won a Nobel Prize for her work to understand why certain cells grow so quickly.
When Rita Levi-Montalcini decided to become a scientist, she was determined that nothing would stand in her way. And from the beginning, that determination was put to the test. Before Levi-Montalcini became a Nobel Prize-winning neurobiologist, the first to discover and isolate a crucial chemical called Neural Growth Factor (NGF), she would have to battle both the sexism within her own family as well as the racism and fascism that was slowly engulfing her country
Levi-Montalcini was born to two loving parents in Turin, Italy at the turn of the 20th century. She and her twin sister, Paola, were the youngest of the family's four children, and Levi-Montalcini described her childhood as "filled with love and reciprocal devotion." But while her parents were loving, supportive and "highly cultured," her father refused to let his three daughters engage in any schooling beyond the basics. "He loved us and had a great respect for women," she later explained, "but he believed that a professional career would interfere with the duties of a wife and mother."
At age 20, Levi-Montalcini had finally had enough. "I realized that I could not possibly adjust to a feminine role as conceived by my father," she is quoted as saying, and asked his permission to finish high school and pursue a career in medicine. When her father reluctantly agreed, Levi-Montalcini was ecstatic: In just under a year, she managed to catch up on her mathematics, graduate high school, and enroll in medical school in Turin.
By 1936, Levi-Montalcini had graduated medical school at the top of her class and decided to stay on at the University of Turin as a research assistant for histologist and human anatomy professor Guiseppe Levi. Levi-Montalcini started studying nerve cells and nerve fibers – the tiny, slender tendrils that are threaded throughout our nerves and that determine what information each nerve can transmit. But it wasn't long before another enormous obstacle to her scientific career reared its head.
Science Under a Fascist Regime
Two years into her research assistant position, Levi-Montalcini was fired, along with every other "non-Aryan Italian" who held an academic or professional career, thanks to a series of antisemitic laws passed by Italy's then-leader Benito Mussolini. Forced out of her academic position, Levi-Montalcini went to Belgium for a fellowship at a neurological institute in Brussels – but then was forced back to Turin when the German army invaded.
Levi-Montalcini decided to keep researching. She and Guiseppe Levi built a makeshift lab in Levi-Montalcini's apartment, borrowing chicken eggs from local farmers and using sewing needles to dissect them. By dissecting the chicken embryos from her bedroom laboratory, she was able to see how nerve fibers formed and died. The two continued this research until they were interrupted again – this time, by British air raids. Levi-Montalcini fled to a country cottage to continue her research, and then two years later was forced into hiding when the German army invaded Italy. Levi-Montalcini and her family assumed different identities and lived with non-Jewish friends in Florence to survive the Holocaust. Despite all of this, Levi-Montalcini continued her work, dissecting chicken embryos from her hiding place until the end of the war.
"The discovery of NGF really changed the world in which we live, because now we knew that cells talk to other cells, and that they use soluble factors. It was hugely important."
A Post-War Discovery
Several years after the war, when Levi-Montalcini was once again working at the University of Turin, a German embryologist named Viktor Hamburger invited her to Washington University in St. Louis. Hamburger was impressed by Levi-Montalcini's research with her chicken embryos, and secured an opportunity for her to continue her work in America. The invitation would "change the course of my life," Levi-Montalcini would later recall.
During her fellowship, Montalcini grew tumors in mice and then transferred them to chick embryos in order to see how it would affect the chickens. To her surprise, she noticed that introducing the tumor samples would cause nerve fibers to grow rapidly. From this, Levi-Montalcini discovered and was able to isolate a protein that she determined was able to cause this rapid growth. She later named this Nerve Growth Factor, or NGF.
From there, Levi-Montalcini and her team launched new experiments to test NGF, injecting it and repressing it to see the effect it had in a test subject's body. When the team injected NGF into embryonic mice, they observed nerve growth, as well as the mouse pups developing faster – their eyes opening earlier and their teeth coming in sooner – than the untreated group. When the team purified the NGF extract, however, it had no effect, leading the team to believe that something else in the crude extract of NGF was influencing the growth of the newborn mice. Stanley Cohen, Levi-Montalcini's colleague, identified another growth factor called EGF – epidermal growth factor – that caused the mouse pups' eyes and teeth to grow so quickly.
Levi-Montalcini continued to experiment with NGF for the next several decades at Washington University, illuminating how NGF works in our body. When Levi-Montalcini injected newborn mice with an antiserum for NGF, for example, her team found that it "almost completely deprived the animals of a sympathetic nervous system." Other experiments done by Levi-Montalcini and her colleagues helped show the role that NGF plays in other important biological processes, such as the regulation of our immune system and ovulation.
"The discovery of NGF really changed the world in which we live, because now we knew that cells talk to other cells, and that they use soluble factors. It was hugely important," said Bill Mobley, Chair of the Department of Neurosciences at the University of California, San Diego School of Medicine.
Her Lasting Legacy
After years of setbacks, Levi-Montalcini's groundbreaking work was recognized in 1986, when she was awarded the Nobel Prize in Medicine for her discovery of NGF (Cohen, her colleague who discovered EGF, shared the prize). Researchers continue to study NGF even to this day, and the continued research has been able to increase our understanding of diseases like HIV and Alzheimer's.
Levi-Montalcini never stopped researching either: In January 2012, at the age of 102, Levi-Montalcini published her last research paper in the journal PNAS, making her the oldest member of the National Academy of Science to do so. Before she died in December 2012, she encouraged other scientists who would suffer setbacks in their careers to keep pursuing their passions. "Don't fear the difficult moments," Levi-Montalcini is quoted as saying. "The best comes from them."

