New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
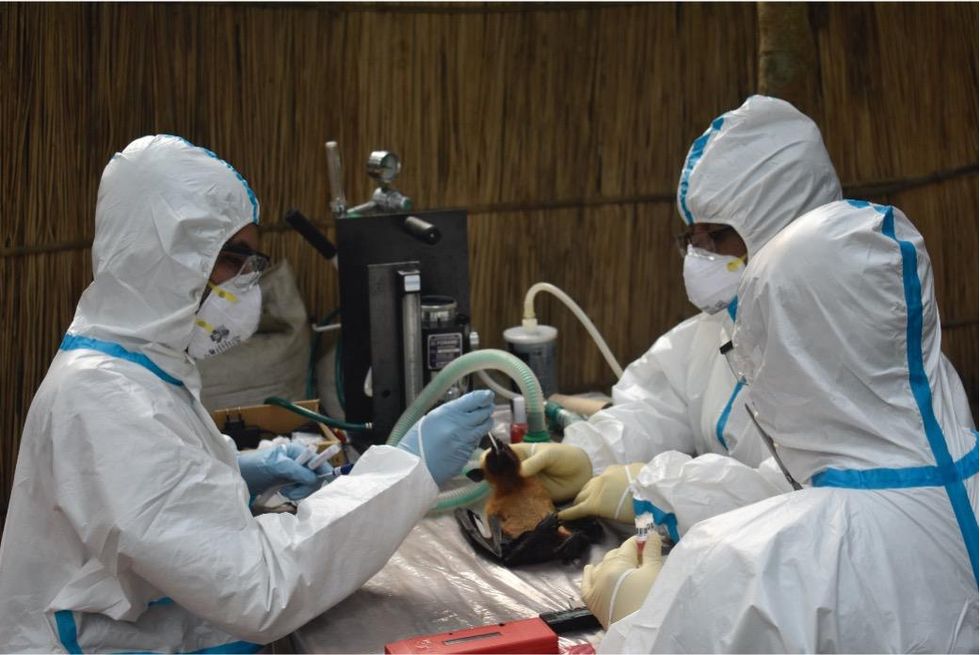
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
Schmidt Ocean Institute co-founder Wendy Schmidt is backed by 32 screens in research vessel Falkor's control room where most of the science takes place on the ship, from mapping to live streaming of underwater robotic dives.
WENDY SCHMIDT is a philanthropist and investor who has spent more than a dozen years creating innovative non-profit organizations to solve pressing global environmental and human rights issues. Recognizing the human dependence on sustaining and protecting our planet and its people, Wendy has built organizations that work to educate and advance an understanding of the critical interconnectivity between the land and the sea. Through a combination of grants and investments, Wendy's philanthropic work supports research and science, community organizations, promising leaders, and the development of innovative technologies. Wendy is president of The Schmidt Family Foundation, which she co-founded with her husband Eric in 2006. They also co-founded Schmidt Ocean Institute and Schmidt Futures.
Editors: The pandemic has altered the course of human history and the nature of our daily lives in equal measure. How has it affected the focus of your philanthropy across your organizations? Have any aspects of the crisis in particular been especially galvanizing as you considered where to concentrate your efforts?
Wendy: The COVID-19 pandemic has made the work of our philanthropy more relevant than ever. If anything, the circumstances of this time have validated the focus we have had for nearly 15 years. We support the need for universal access to clean, renewable energy, healthy food systems, and the dignity of human labor and self-determination in a world of interconnected living systems on land and in the Ocean we are only beginning to understand.
When you consider the disproportionate impact of the COVID-19 virus on people who are poorly paid, poorly housed, with poor nutrition and health care, and exposed to unsafe conditions in the workplace—you see clearly how the systems that have been defining how we live, what we eat, who gets healthcare and what impacts the environment around us—need to change.
"This moment has propelled broad movements toward open publication and open sharing of data and samples—something that has always been a core belief in how we support and advance science."
If the pandemic teaches us anything, we learn what resilience looks like, and the essential role for local small businesses including restaurants, farms and ranches, dairies and fish markets in the long term vitality of communities. There is resonance, local economic benefit, and also accountability in these smaller systems, with shorter supply chains and less vertical integration.
The consolidation of vertically integrated business operations for the sake of global efficiency reveals its essential weakness when supply chains break down and the failure to encourage local economic centers leads to intense systemic disruption and the possibility of collapse.
Editors: For scientists, one significant challenge has been figuring out how to continue research, if at all, during this time of isolation and distancing. Yet, your research vessel Falkor, of the Schmidt Ocean Institute, is still on its expedition exploring the Coral Sea Marine Park in Australia—except now there are no scientists onboard. What was the vessel up to before the pandemic hit? Can you tell us more about how they are continuing to conduct research from afar now and how that's going?
Wendy: We have been extremely fortunate at Schmidt Ocean Institute. When the pandemic hit in March, our research vessel, Falkor, was already months into a year-long program to research unexplored deep sea canyons around Australia and at the Great Barrier Reef. We were at sea, with an Australian science group aboard, carrying on with our mission of exploration, discovery and communication, when we happened upon what we believe to be the world's longest animal—a siphonophore about 150 feet long, spiraling out at a depth of about 2100 feet at the end of a deeper dive in the Ningaloo Canyon off Western Australia. It was the kind of wondrous creature we find so often when we conduct ROV dives in the world's Ocean.
For more than two months this year, Falkor was reportedly the only research vessel in the world carrying on active research at sea. Once we were able to dock and return the science party to shore, we resumed our program at sea offering a scheduled set of now land-based scientists in lockdown in Australia the opportunity to conduct research remotely, taking advantage of the vessel's ship to shore communications, high resolution cameras and live streaming video. It's a whole new world, and quite wonderful in its own way.
Editors: Normally, 10–15 scientists would be aboard such a vessel. Is "remote research" via advanced video technology here to stay? Are there any upsides to this "new normal"?
Wendy: Like all things pandemic, remote research is an adaptation for what would normally occur. Since we are putting safety of the crew and guest scientists at the forefront, we're working to build strong remote connections between our crew, land based scientists and the many robotic tools on board Falkor. There's no substitute for in person work, but what we've developed during the current cruise is a pretty good and productive alternative in a crisis. And what's important is that this critical scientific research into the deep sea is able to continue, despite the pandemic on land.
Editors: Speaking of marine expeditions, you've sponsored two XPRIZE competitions focused on ocean health. Do you think challenge prizes could fill gaps of the global COVID-19 response, for example, to manufacture more testing kits, accelerate the delivery of PPE, or incentivize other areas of need?
Wendy: One challenge we are currently facing is that innovations don't have the funding pathway to scale, so promising ideas by entrepreneurs, researchers, and even major companies are being developed too slowly. Challenge prizes help raise awareness for problems we are trying to solve and attract new people to help solve those problems by giving them a pathway to contribute.
One idea might be for philanthropy to pair prizes and challenges with an "advanced market commitment" where the government commits to a purchase order for the innovation if it meets a certain test. That could be deeply impactful for areas like PPE and the production of testing kits.
Editors: COVID-19 testing, especially, has been sorely needed, here in the U.S. and in developing countries as well as low-income communities. That's why we're so intrigued by your Schmidt Science Fellows grantee Hal Holmes and his work to repurpose a new DNA technology to create a portable, mobile test for COVID-19. Can you tell us about that work and how you are supporting it?
Wendy: Our work with Conservation X Labs began years ago when our foundation was the first to support their efforts to develop a handheld DNA barcode sensor to help detect illegally imported and mislabeled seafood and timber products. The device was developed by Hal Holmes, who became one of our Schmidt Science Fellows and is the technical lead on the project, working closely with Conservation X Labs co-founders Alex Deghan and Paul Bunje. Now, with COVID-19, Hal and team have worked with another Schmidt Science Fellow, Fahim Farzardfard, to repurpose the technology—which requires no continuous power source, special training, or a lab—to serve as a mobile testing device for the virus.
The work is going very well, manufacturing is being organized, and distribution agreements with hospitals and government agencies are underway. You could see this device in use within a few months and have testing results within hours instead of days. It could be especially useful in low-income communities and developing countries where access to testing is challenging.
Editors: How is Schmidt Futures involved in the development of information platforms that will offer productive solutions?
Wendy: In addition to the work I've mentioned, we've also funded the development of tech-enabled tools that can help the medical community be better prepared for the ongoing spike of COVID cases. For example, we funded EdX and Learning Agency to develop an online training to help increase the number of medical professionals who can operate ventilators. The first course is being offered by Harvard University, and so far, over 220,000 medical professionals have enrolled. We have also invested in informational platforms that make it easier to contain the spread of the disease, such as our work with Recidiviz to model the impact of COVID-19 in prisons and outline policy steps states could take to limit the spread.
Information platforms can also play a big part pushing forward scientific research into the virus. For example, we've funded the UC Santa Cruz Virus Browser, which allows researchers to examine each piece of the virus and see the proteins it creates, the interactions in the host cell, and — most importantly — almost everything the recent scientific literature has to say about that stretch of the molecule.
Editors: The scale of research collaboration and the speed of innovation today seem unprecedented. The whole science world has turned its attention to combating the pandemic. What positive big-picture trends do you think or hope will persist once the crisis eventually abates?
Wendy: As in many areas, the COVID crisis has accelerated trends in the scientific world that were already well underway. For instance, this moment has propelled broad movements toward open publication and open sharing of data and samples—something that has always been a core belief in how we support and advance science.
We believe collaboration is an essential ingredient for progress in all areas. Early in this pandemic, Schmidt Futures held a virtual gathering of 160 people across 70 organizations in philanthropy, government, and business interested in accelerating research and response to the virus, and thought at the time, it's pretty amazing this kind of thing doesn't go all the time. We are obviously going to go farther together than on our own...
My husband, Eric, has observed that in the past two months, we've all catapulted 10 years forward in our use of technology, so there are trends already underway that are likely accelerated and will become part of the fabric of the post-COVID world—like working remotely; online learning; increased online shopping, even for groceries; telemedicine; increasing use of AI to create smarter delivery systems for healthcare and many other applications in a world that has grown more virtual overnight.
"Our deepest hope is that out of these alarming and uncertain times will come a renewed appreciation for the tools of science, as they help humans to navigate a world of interconnected living systems, of which viruses are a large part."
We fully expect these trends to continue and expand across the sciences, sped up by the pressures of the health crisis. Schmidt Ocean Institute and Schmidt Futures have been pressing in these directions for years, so we are pleased to see the expansions that should help more scientists work productively, together.
Editors: Trying to find the good amid a horrible crisis, are there any other new horizons in science, philanthropy, and/or your own work that could transform our world for the better that you'd like to share?
Wendy: Our deepest hope is that out of these alarming and uncertain times will come a renewed appreciation for the tools of science, as they help humans to navigate a world of interconnected living systems, of which viruses are a large part. The more we investigate the Ocean, the more we look deeply into what lies in our soils and beneath them, the more we realize we do not know, and moreover, how vulnerable humanity is to the forces of the natural world.
Philanthropy has an important role to play in influencing how people perceive our place in the world and understand the impact of human activity on the rest of the planet. I believe it's philanthropy's role to take risks, to invest early in innovative technologies, to lead where governments and industry aren't ready to go yet. We're fortunate at this time to be able to help those working on tools to better diagnose and treat the virus, and to invest in those working to improve information systems, so citizens and policy makers can make better decisions that can reduce impacts on families and institutions.
From all we know, this isn't likely to be the last pandemic the world will see. It's been said that a crisis comes before change, and we would hope that we can play a role in furthering the work to build systems that are resilient—in information, energy, agriculture and in all the ways we work, recreate, and use the precious resources of our planet.
[This article was originally published on June 8th, 2020 as part of a standalone magazine called GOOD10: The Pandemic Issue. Produced as a partnership among LeapsMag, The Aspen Institute, and GOOD, the magazine is available for free online.]
Kira Peikoff was the editor-in-chief of Leaps.org from 2017 to 2021. As a journalist, her work has appeared in The New York Times, Newsweek, Nautilus, Popular Mechanics, The New York Academy of Sciences, and other outlets. She is also the author of four suspense novels that explore controversial issues arising from scientific innovation: Living Proof, No Time to Die, Die Again Tomorrow, and Mother Knows Best. Peikoff holds a B.A. in Journalism from New York University and an M.S. in Bioethics from Columbia University. She lives in New Jersey with her husband and two young sons. Follow her on Twitter @KiraPeikoff.
Dr. Barry Marshall (along with a collaborator) discovered that the bacteria H. Pylori, pictured, caused stomach ulcers.
[Editor's Note: Welcome to Leaps of the Past, a new monthly column that spotlights the fascinating backstory behind a medical or scientific breakthrough from history.]
------
Until about 40 years ago, ulcers were a mysterious – and sometimes deadly – ailment. Found in a person's stomach lining or intestine, ulcers are small sores that cause a variety of painful symptoms, such as vomiting, a burning or aching sensation, internal bleeding and stomach obstruction. Patients with ulcers suffered for years without a cure and sometimes even needed their stomachs completely removed to rid them from pain.
"To gastroenterologists, the concept of a germ causing ulcers was like saying the Earth is flat."
In the early 1980s, the majority of scientists thought that ulcers were caused by stress or poor diet. But a handful of scientists had a different theory: They believed that ulcers were caused by a corkscrew-shaped bacterium called Helicobacter pylori, or H. pylori for short. Robin Warren, a pathologist, and Barry Marshall, an internist, were the two pioneers of this theory, and the two teamed up to study H. pylori at the Royal Perth Hospital in 1981.
The pair started off by trying to culture the bacteria in the stomachs of patients with gastritis, an inflammation of the stomach lining and a precursor to developing an ulcer. Initially, the microbiologists involved in their clinical trial found no trace of the bacteria from patient samples – but after a few weeks, the microbiologists discovered that their lab techs had been throwing away the cultures before H. pylori could grow. "After that, we let the cultures grow longer and found 13 patients with duodenal ulcer," said Marshall in a later interview. "All of them had the bacteria."
Marshall and Warren also cultured H. pylori in the stomachs of patients with stomach cancer. They observed that "everybody with stomach cancer developed it on a background of gastritis. Whenever we found a person without Helicobacter, we couldn't find gastritis either." Marshall and Warren were convinced that H. pylori not only caused gastritis and peptic ulcers, but stomach cancer as well.
But when the team presented their findings at an annual meeting of the Royal Australasian College of Physicians in Perth, they were mostly met with skepticism. "To gastroenterologists, the concept of a germ causing ulcers was like saying the Earth is flat," Marshall said. "The idea was too weird."
Warren started treating his gastritis patients with antibiotics with great success – but other internists remained doubtful, continuing to treat their patients with antacids instead. Making matters more complicated, neither Warren nor Marshall could readily test their theory, since the pair only had lab mice at their disposal and H. pylori infects only humans and non-human primates, such as rhesus monkeys.
So Marshall took an unconventional approach. First, he underwent two tests to get a baseline reading of his stomach, which showed no presence of H. pylori. Then, Marshall took some H. pylori bacteria from a petri dish, mixed it with beef extract to create a broth, and gulped it down. If his theory was correct, a second gastric biopsy would show that his stomach was overrun with H. pylori bacteria, and a second endoscopy would show a painfully inflamed stomach – gastritis.
Less than a week later, Marshall started feeling sick. "I expected to develop an asymptomatic infection," he later said in an interview published in the Canadian Journal of Gastroenterology. "… [but] after five days, I started to have bloating and fullness after the evening meal, and my appetite decreased. My breath was bad and I vomited clear watery liquid, without acid, each morning."
At his wife's urging, Marshall started on a regimen of antibiotics to kill off the burgeoning bacteria, so a follow-up biopsy showed no signs of H. pylori. A follow-up endoscopy, however, showed "severe active gastritis" along with epithelial damage. This was the smoking gun other clinicians needed to believe that H. pylori caused gastritis and stomach cancer. When they began to treat their gastritis patients with antibiotics, the rate of peptic ulcers in the Australian population diminished by 70 percent.
Today, antibiotics are the standard of care for anyone afflicted with gastritis.
In 2005, Marshall and Warren were awarded the Nobel Prize in Physiology or Medicine for their discovery of H. Pylori and its role in developing gastritis and peptic ulcers. "Thanks to the pioneering discovery by Marshall and Warren, peptic ulcer disease is no longer a chronic, frequently disabling condition, but a disease that can be cured by a short regimen of antibiotics and acid secretion inhibitors," the Nobel Prize Committee said.
Today, antibiotics are the standard of care for anyone afflicted with gastritis – and stomach cancer has been significantly reduced in the Western world.

