New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
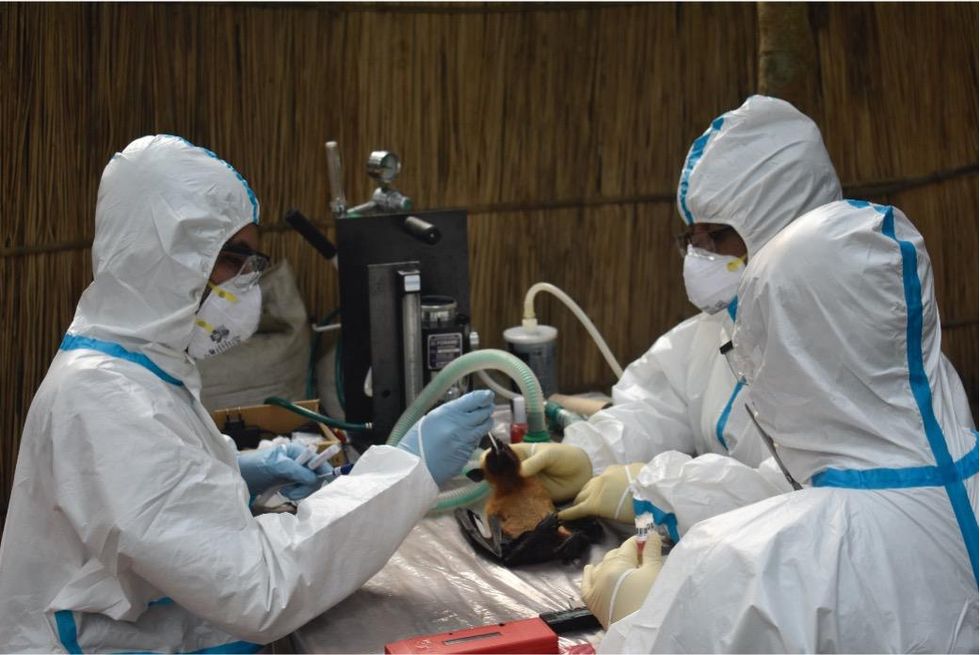
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
The latest tools, like whole genome sequencing, are allowing food outbreaks to be identified and stopped more quickly.
With the pandemic at the forefront of everyone's minds, many people have wondered if food could be a source of coronavirus transmission. Luckily, that "seems unlikely," according to the CDC, but foodborne illnesses do still sicken a whopping 48 million people per year.
Whole genome sequencing is like "going from an eight-bit image—maybe like what you would see in Minecraft—to a high definition image."
In normal times, when there isn't a historic global health crisis infecting millions and affecting the lives of billions, foodborne outbreaks are real and frightening, potentially deadly, and can cause widespread fear of particular foods. Think of Romaine lettuce spreading E. coli last year— an outbreak that infected more than 500 people and killed eight—or peanut butter spreading salmonella in 2008, which infected 167 people.
The technologies available to detect and prevent the next foodborne disease outbreak have improved greatly over the past 30-plus years, particularly during the past decade, and better, more nimble technologies are being developed, according to experts in government, academia, and private industry. The key to advancing detection of harmful foodborne pathogens, they say, is increasing speed and portability of detection, and the precision of that detection.
Getting to Rapid Results
Researchers at Purdue University have recently developed a lateral flow assay that, with the help of a laser, can detect toxins and pathogenic E. coli. Lateral flow assays are cheap and easy to use; a good example is a home pregnancy test. You place a liquid or liquefied sample on a piece of paper designed to detect a single substance and soon after you get the results in the form of a colored line: yes or no.
"They're a great portable tool for us for food contaminant detection," says Carmen Gondhalekar, a fifth-year biomedical engineering graduate student at Purdue. "But one of the areas where paper-based lateral flow assays could use improvement is in multiplexing capability and their sensitivity."
J. Paul Robinson, a professor in Purdue's Colleges of Veterinary Medicine and Engineering, and Gondhalekar's advisor, agrees. "One of the fundamental problems that we have in detection is that it is hard to identify pathogens in complex samples," he says.
When it comes to foodborne disease outbreaks, you don't always know what substance you're looking for, so an assay made to detect only a single substance isn't always effective. The goal of the project at Purdue is to make assays that can detect multiple substances at once.
These assays would be more complex than a pregnancy test. As detailed in Gondhalekar's recent paper, a laser pulse helps create a spectral signal from the sample on the assay paper, and the spectral signal is then used to determine if any unique wavelengths associated with one of several toxins or pathogens are present in the sample. Though the handheld technology has yet to be built, the idea is that the results would be given on the spot. So someone in the field trying to track the source of a Salmonella infection could, for instance, put a suspected lettuce sample on the assay and see if it has the pathogen on it.
"What our technology is designed to do is to give you a rapid assessment of the sample," says Robinson. "The goal here is speed."
Seeing the Pathogen in "High-Def"
"One in six Americans will get a foodborne illness every year," according to Dr. Heather Carleton, a microbiologist at the Centers for Disease Control and Prevention's Enteric Diseases Laboratory Branch. But not every foodborne outbreak makes the news. In 2017 alone, the CDC monitored between 18 and 37 foodborne poison clusters per week and investigated 200 multi-state clusters. Hardboiled eggs, ground beef, chopped salad kits, raw oysters, frozen tuna, and pre-cut melon are just a taste of the foods that were investigated last year for different strains of listeria, salmonella, and E. coli.
At the heart of the CDC investigations is PulseNet, a national network of laboratories that uses DNA fingerprinting to detect outbreaks at local and regional levels. This is how it works: When a patient gets sick—with symptoms like vomiting and fever, for instance—they will go to a hospital or clinic for treatment. Since we're talking about foodborne illnesses, a clinician will likely take a stool sample from the patient and send it off to a laboratory to see if there is a foodborne pathogen, like salmonella, E. Coli, or another one. If it does contain a potentially harmful pathogen, then a bacterial isolate of that identified sample is sent to a regional public health lab so that whole genome sequencing can be performed.
Whole genome sequencing can differentiate "virtually all" strains of foodborne pathogens, no matter the species, according to the FDA.
Whole genome sequencing is a method for reading the entire genome of a bacterial isolate (or from any organism, for that matter). Instead of working with a couple dozen data points, now you're working with millions of base pairs. Carleton likes to describe it as "going from an eight-bit image—maybe like what you would see in Minecraft—to a high definition image," she says. "It's really an evolution of how we detect foodborne illnesses and identify outbreaks."
If the bacterial isolate matches another in the CDC's database, this means there could be a potential outbreak and an investigation may be started, with the goal of tracking the pathogen to its source.
Whole genome sequencing has been a relatively recent shift in foodborne disease detection. For more than 20 years, the standard technique for analyzing pathogens in foodborne disease outbreaks was pulsed-field gel electrophoresis. This method creates a DNA fingerprint for each sample in the form of a pattern of about 15-30 "bands," with each band representing a piece of DNA. Researchers like Carleton can use this fingerprint to see if two samples are from the same bacteria. The problem is that 15-30 bands are not enough to differentiate all isolates. Some isolates whose bands look very similar may actually come from different sources and some whose bands look different may be from the same source. But if you can see the entire DNA fingerprint, then you don't have that issue. That's where whole genome sequencing comes in.
Although the PulseNet team had piloted whole genome sequencing as early as 2013, it wasn't until July of last year that the transition to using whole genome sequencing for all pathogens was complete. Though whole genome sequencing requires far more computing power to generate, analyze, and compare those millions of data points, the payoff is huge.
Stopping Outbreaks Sooner
The U.S. Food and Drug Administration (FDA) acquired their first whole genome sequencers in 2008, according to Dr. Eric Brown, the Director of the Division of Microbiology in the FDA's Office of Regulatory Science. Since then, through their GenomeTrakr program, a network of more than 60 domestic and international labs, the FDA has sequenced and publicly shared more than 400,000 isolates. "The impact of what whole genome sequencing could do to resolve a foodborne outbreak event was no less impactful than when NASA turned on the Hubble Telescope for the first time," says Brown.
Whole genome sequencing has helped identify strains of Salmonella that prior methods were unable to differentiate. In fact, whole genome sequencing can differentiate "virtually all" strains of foodborne pathogens, no matter the species, according to the FDA. This means it takes fewer clinical cases—fewer sick people—to detect and end an outbreak.
And perhaps the largest benefit of whole genome sequencing is that these detailed sequences—the millions of base pairs—can imply geographic location. The genomic information of bacterial strains can be different depending on the area of the country, helping these public health agencies eventually track the source of outbreaks—a restaurant, a farm, a food-processing center.
Coming Soon: "Lab in a Backpack"
Now that whole genome sequencing has become the go-to technology of choice for analyzing foodborne pathogens, the next step is making the process nimbler and more portable. Putting "the lab in a backpack," as Brown says.
The CDC's Carleton agrees. "Right now, the sequencer we use is a fairly big box that weighs about 60 pounds," she says. "We can't take it into the field."
A company called Oxford Nanopore Technologies is developing handheld sequencers. Their devices are meant to "enable the sequencing of anything by anyone anywhere," according to Dan Turner, the VP of Applications at Oxford Nanopore.
"The sooner that we can see linkages…the sooner the FDA gets in action to mitigate the problem and put in some kind of preventative control."
"Right now, sequencing is very much something that is done by people in white coats in laboratories that are set up for that purpose," says Turner. Oxford Nanopore would like to create a new, democratized paradigm.
The FDA is currently testing these types of portable sequencers. "We're very excited about it. We've done some pilots, to be able to do that sequencing in the field. To actually do it at a pond, at a river, at a canal. To do it on site right there," says Brown. "This, of course, is huge because it means we can have real-time sequencing capability to stay in step with an actual laboratory investigation in the field."
"The timeliness of this information is critical," says Marc Allard, a senior biomedical research officer and Brown's colleague at the FDA. "The sooner that we can see linkages…the sooner the FDA gets in action to mitigate the problem and put in some kind of preventative control."
At the moment, the world is rightly focused on COVID-19. But as the danger of one virus subsides, it's only a matter of time before another pathogen strikes. Hopefully, with new and advancing technology like whole genome sequencing, we can stop the next deadly outbreak before it really gets going.
What Will Make the Public Trust a COVID-19 Vaccine?
A successful deployment of an eventual vaccine will mean grappling with ongoing cultural tensions.
With a brighter future hanging on the hopes of an approved COVID-19 vaccine, is it possible to win over the minds of fearful citizens who challenge the value or safety of vaccination?
Globally, nine COVID-19 vaccines so far are being tested for safety in early phase human clinical trials.
It's a decades-old practice. With a dose injected into the arm of a healthy patient, doctors aim to prevent illness with a vaccine shot designed to trigger a person's immune system to fight serious infection without getting the disease.
This week, in fact, the U.S. frontrunner vaccine candidate, developed by Moderna, safely produced an immune response in the first eight healthy volunteers, the company announced. A large efficacy trial is planned to start in July. But if positive signals for safety and efficacy result from that trial, will that be enough to convince the public to broadly embrace a new vaccine?
"Throughout the history of vaccines there has always been a small vocal minority who don't believe vaccines work or don't trust the science," says sociologist and researcher Jennifer Reich, a professor at the University of Colorado in Denver and author of Calling the Shots: Why Parents Reject Vaccines.
Research indicates that only about 2 percent of the population say vaccines aren't necessary under any circumstance. Remarkably, a quarter to one third of American parents delay or reject the shots, not because they are anti-vaccine, but because they disapprove of the recommended timing or administration, says Reich.
Additionally, addressing distrust about how they come to market is key when talking to parents, workers or anyone targeted for a new vaccine, she says.
"When I talk to parents about why they reject vaccines for their kids, a lot of them say that they don't fully trust the process by which vaccines are regulated and tested," says Reich. "They don't trust that vaccine manufacturers -- which are for-profit companies -- are looking out for public health."
Balancing Act
Globally, nine COVID-19 vaccine candidates so far are being tested for safety in early phase human clinical trials and more than 100 are under development as scientists hustle to curtail the disease. Creating a new vaccine at a record pace requires a delicate balance of benefit and risk, says vaccinology expert Dr. Kathryn Edwards, professor of pediatrics in the division of infectious diseases at Vanderbilt University School of Medicine in Nashville, Tenn.
"We take safety very seriously," says Dr. Edwards. "We don't want something bad to happen, but we also realize that we have a terrible outbreak and we have a lot of people dying. We want to figure out how we can stop this."
In the U.S., all vaccine clinical trials have a data safety board of experts who monitor results for adverse reactions and red flags that should halt a study, notes Dr. Edwards. Any candidate that succeeds through safety and efficacy trials still requires review and approval by the Food and Drug Administration before a public launch.
Community vs. Individual
A major challenge to the deployment of a safe and effective coronavirus vaccine goes beyond the technical realm. A persistent all-out anti-vaccine sentiment has found a home and growing community on social media where conspiracies thrive. Main tenets of the movement are that vaccines are ineffective, unsafe and cause autism, despite abundant scientific evidence to the contrary.
Best-case scenario, more than one successful vaccine ascends with competing methods to achieve the same goal of preventing or lessening the severity of the COVID-19 virus.
In fact, widespread use of vaccines is considered by the U.S. Centers of Disease Control and Prevention to be one of the greatest public health achievements of the 20th Century. The World Health Organization estimates that between two million to three million deaths are avoided each year through immunization campaigns that employ vaccination to control life-threatening infectious diseases.
Most people reluctant to give their children vaccines, however, don't oppose them for everyone, but believe that they are a personal choice, says Reich.
"They think that vaccines are one strategy in personal health optimization, but they shouldn't be mandated for participation in any part of civil society," she says.
Vaccine hesitancy, like the teeter totter of social distancing acceptance, reflects the push and pull of individual versus community values, says Reich.
"A lot of people are saying, 'I take personal responsibility for my own health and I don't want a city or a county or state telling me what I should and shouldn't do,'" says Reich. "Then we also see calls for collective responsibility that says 'It's not your personal choice. This is about helping health systems function. This is about making sure vulnerable people are protected.'"
These same debates are likely to continue if a vaccine comes to market, she says.
Building Public Confidence
Reich offers solutions to address the conflict between embedded American norms and widespread embrace of an approved COVID-19 vaccine. Long-term goals: Stop blaming people when they get sick, treat illness as a community responsibility, make sick leave common for all workers, and improve public health systems.
"In the shorter run," says Reich, "health authorities and companies that might bring a vaccine to market need to work very hard to explain to the public why they should trust this vaccine and why they should use it."
The rush for a viable vaccine raises questions for consumers. To build public confidence, it's up to FDA reviewers, institutions and pharmaceutical companies to explain "what steps were skipped. What steps moved forward. How rigorous was safety testing. And to make that information clear to the public," says Reich.
Dr. Edwards says clinical trial timelines accelerated to test vaccines in humans make all the safeguards involved in the process that more compelling and important.
"There's no question we need a vaccine," she says. "But we also have to make sure that we don't harm people."
The Road Ahead
Think of manufacturing and distribution as key pitstops to keep the race for a vaccine on the road to the finish line. Both elements require substantial effort and consideration.
The speed of getting a vaccine to those who need it could hinge on the type of technology used to create it. Best-case scenario, more than one successful vaccine ascends with competing methods to achieve the same goal of preventing or lessening the severity of the COVID-19 virus.
Technological platforms fall into two basic camps, those that are proven and licensed for other viruses, and experimental approaches that may hold great promise but lack regulatory approval, says Maria Elena Bottazzi, co-director of Texas Children's Center for Vaccine Development at Baylor College of Medicine in Houston.
Moderna, for instance, employs an experimental technology called messenger RNA (mRNA) that has produced the encouraging early results in human safety trials, although some researchers criticized the company for not making the data public. The mRNA vaccine instructs cells to make copies of the key COVID-19 spike protein, with the goal of then triggering production of immune cells that can recognize and attack the virus if it ever invades the body.
"We were already seeing a lot of dissent around questions of individual freedoms and community responsibilities."
Scientists always look for ways to incorporate new technologies into drug development, says Bottazzi. On the other hand, the more basic and generic the technology, theoretically, the faster production could ramp up if a vaccine proves successful through all phases of clinical trials, she says.
"I don't want to develop a vaccine in my lab, but then I don't have anybody to hand it off to because my process is not suitable" for manufacturing or scalability, says Bottazzi.
Researchers at the Baylor lab hope to repurpose a shelved vaccine developed for the genetically similar SARS virus, with a strategy to leverage what is already known instead of "starting from scratch" to develop a COVID-19 vaccine. A recombinant protein technology similar to that used for an approved Hepatitis B vaccine lets scientists focus on identifying a suitable vaccine target without the added worry of a novel platform, says Bottazzi.
The Finish Line
If and when a COVID-19 vaccine is approved is anyone's guess. Announcing a plan to hasten vaccine development via a program dubbed Operation Warp Speed, President Trump said recently one could be available "hopefully" by the end of the year or early 2021.
Scientists urge caution, noting that safe vaccines can take 10 years or more to develop. If a rushed vaccine turns out to have safety and efficacy issues, that could add ammunition to the anti-vaccine lobby.
Emergence of a successful vaccine requires an "enormous effort" with many complex systems from the lab all the way to manufacturing enough capacity to handle a pandemic, says Bottazzi.
"At the same time, you're developing it, you're really carefully assessing its safety and ability to be effective," she says, so it's important "not to get discouraged" if it takes longer than a year or more.
To gauge if a vaccine works on a broad scale, it would have to be delivered into communities where the virus is active. There are examples in history of life-saving vaccines going first to people who could pay for them and not to those who needed them most, says Reich.
"Agencies are going to have to think about how those distribution decisions are going to be made and who is going to make them and that will go a certain way toward reassuring the public," says Reich.
A Gallup survey last year found that vaccine confidence, in general, remains high, with 86 percent of Americans believing that vaccines are safer than the diseases that they are designed to prevent. Still, recent news organization polls indicate that roughly 20 to 25 percent of Americans say they won't or are unlikely to get a COVID-19 vaccine if one becomes available.
Until the 1980s, every vaccine to hit the market was appreciated; a culture of questioning science didn't exist in the same way as today, notes Reich. Time passed and attitudes changed.
"We were already having robust arguments nationally about what counts as an expert, what's the role of the government in daily life," says Reich. "We were already seeing a lot of dissent around questions of individual freedoms and community responsibilities. COVID-19 did not create those conflicts, but they've definitely become more visible since we've moved into this pandemic."

