New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
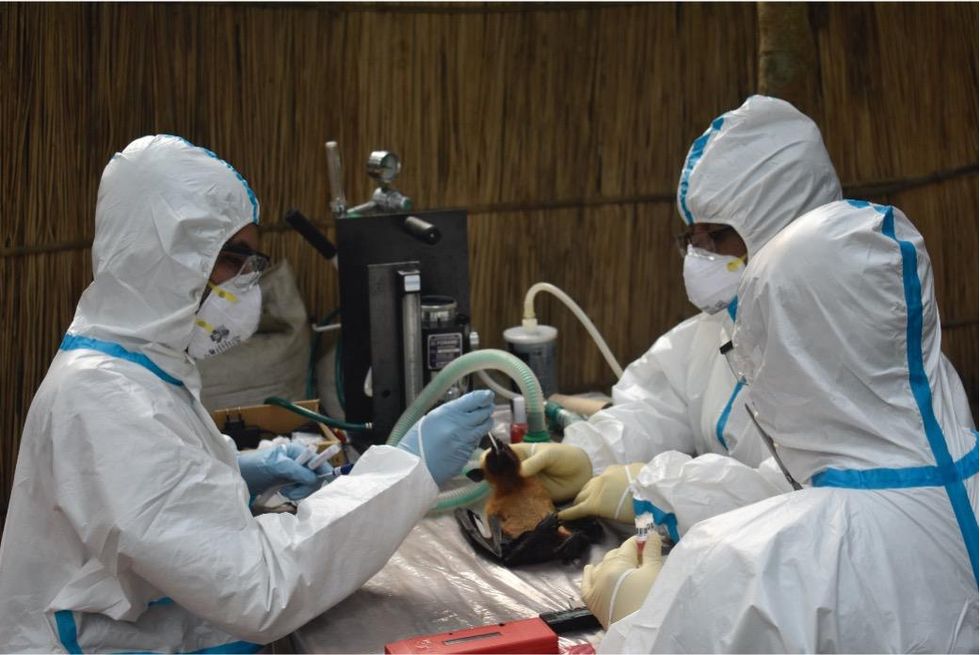
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
Move Over, Iron Man. A Real-Life Power Suit Helped This Paralyzed Grandmother Learn to Run.
Puschel Sorensen undergoing physical therapy using Cyberdyne's hybrid assistive limb, left, and with her grandson while still in a wheelchair.
Puschel Sorensen first noticed something was wrong when her fingertips began to tingle. Later that day, she grew weak and fell.
It picked up small electrical impulses on her skin's surface and turned them into full movement in her legs.
Her family rushed her to the doctor, where she received the devastating diagnosis of Guillain-Barré Syndrome -- a rare and rapidly progressing autoimmune disorder that attacks the myelin sheath covering nerves.
Sorensen, a once-spry grandmother in her late fifties, spent 54 days in intensive care in 2018. When she was finally transferred to a rehab facility near her home in Florida, she was still on a feeding tube and ventilator, and was paralyzed from the neck down. Progress with traditional physical therapy was slow.

Sorensen in the hospital after her diagnosis of Guillain-Barré syndrome.
And then everything changed. Sorensen began using a cutting-edge technology called an exoskeleton to relearn how to walk. In the vein of Iron Man's fictional power suit, it confers strength and mobility to the wearer that isn't possible otherwise. In Sorensen's case, her device, called HAL – for hybrid assistive limb -- picked up small electrical impulses on her skin's surface and turned them into full movement in her legs while she attempted to walk on a treadmill.
"It was very difficult, but super awesome," recalls Sorensen, of first using the device. "The robot was having to do all the work for me."
Amazingly, within a year, she was running. She's one of 38 patients who have used HAL to recover from accidents or medical catastrophes.
"How do you thank someone for giving them back the ability to walk, the ability to live your life again?" Sorensen asks effusively.
It's still early days for such exoskeleton devices, which number perhaps a few thousand worldwide, according to data from the handful of manufacturers who create them with any scale. But the devices' ability to dramatically rehabilitate patients like Sorensen highlights their potential to extract untold numbers of people from wheelchairs, and even to usher in a new paradigm for caregiving – one of the fastest growing segments of the U.S. economy.
"I've been a physical therapist for 16 years, and (these devices) help teach patients the right way to move in rehabilitation," says Robert McIver, director of clinical technology at the Brooks Cybernic Treatment Center, part of the Brooks Rehabilitation Hospital in Jacksonville, Fla, where Sorensen recovered.
Another patient there, a 17-year-old named George with a snowboarding injury that paralyzed his legs, was getting around with a walker within 20 sessions.
As patients progress in their recoveries, so does exoskeleton technology. Jack Peurach, CEO of Ekso, one of the leaders in the space, believes within a decade they could resemble an article of clothing (a "magic pair of pants" is his phrase). They also may become inexpensive and reliable enough to transition from a medical to a consumer device. McIver sees them eventually being used in the home on an ongoing basis as a personal assistive device, much like a walker or cane, to prevent falls in elderly people.
Such a transition "certainly could eventually lessen the need for caregivers," says Sharona Hoffman, a professor of law at Case Western University in Cleveland who has written extensively on aging and bioethics. "We have a real shortage of caregivers, so that would be a good thing."
Of course, having an aging and disabled population using exoskeletons in much the same way as an Apple Watch raises issues of its own.
Dr. Elizabeth Landsverk, a California-based geriatrician and founder of a company that performs house calls for elderly patients, believes the tech holds some promise in easing the burden on caregivers, who sometimes have to lift or move patients without assistance. But she also believes exoskeletons could become overhyped.
"I don't see robotics as completely replacing the caregiver," she says. And even if exoskeletons became akin to articles of clothing, she is skeptical of how convenient they could become.
"It's hard enough to get into support hose. Would an older person be able to get in and out of it on their own?" she asks, noting that a patient's cognitive levels could pose a huge barrier to donning such a device without assistance.
If personal exoskeletons did wildly succeed, Hoffman wonders whether they would leave the elderly more physically mobile yet also more socially isolated, since caregivers or even residing in an assisted living facility may no longer be required. Or, if they were priced in the hundreds or thousands of dollars, he worries that the cost would exacerbate social inequalities among the elderly and disabled.
"It's almost like a bad dream that [my illness] happened."
With any technology that confers superhuman ability, there's also the question of appropriate usage. Even the fictional Power Loader in the movie Alien required an operator's license. In the real world, such an approach would likely pay dividends.
"We would have to make sure physicians are well-trained in these devices, and patients have a way of getting training to operate them that is thorough and responsible," Hoffman says.
But despite some unresolved questions, it is a remarkable achievement to be able to give people back their lives thanks to new technology.
"It's almost like a bad dream that [my illness] happened," says Sorensen, who managed to walk in her daughter's wedding after her recovery. "Because now everything is pretty much back to normal and it's awesome."
23andMe Is Using Customers’ Genetic Data to Develop Drugs. Is This Brilliant or Dubious?
A woman does a DNA test with a cotton swab at home.
Leading direct-to-consumer (DTC) genetic testing companies are continuously unveiling novel ways to leverage their vast stores of genetic data.
"23andMe will tell you what diseases you have and then sell you the drugs to treat them."
As reported last week, 23andMe's latest concept is to develop and license new drugs using the data of consumers who have opted in to let their information be used for research. To date, over 10 million people have used the service and around 80 percent have opted in, making its database one of the largest in the world.
Culture researcher Dr. Julia Creet is one of the foremost experts on the DTC genetic testing industry, and in her forthcoming book, The Genealogical Sublime, she bluntly examines whether such companies' motives and interests are in sync with those of consumers.
Leapsmag caught up with Creet about the latest news and the wider industry's implications for health and privacy.
23andMe has just announced that it plans to license a newly developed anti-inflammatory drug, the first one created using its customers' genetic data, to Almirall, a pharma company in Spain. What's your take?
I think this development is the next step in the evolution of the company and its "double-sided" marketing model. In the past, as it enticed customers to give it their DNA, it sold the results and the medical information divulged by customers to other drug companies. Now it is positioning itself to reap the profits of a new model by developing treatments itself.
Given that there are many anti-inflammatory drugs on the market already, whatever Almirall produces might not have much of an impact. We might see this canny move as a "proof of concept," that 23andMe has learned how to "leverage" its genetic data without having to sell them to a third party. In a way, the privacy provisions will be much less complicated, and the company stands to attract investment as it turns itself into [a pseudo pharmaceutical company], a "pharma-psuedocal" company.
Emily Drabant Conley, the president of business development, has said that 23andMe is pursuing other drug compounds and may conduct their own clinical trials rather than licensing them out to their existing research partners. The end goal, it seems, is to make direct-to-consumer DNA testing to drug production and sales back to that same consumer base a seamless and lucrative circle. You have to admit it's a brilliant business model. 23andMe will tell you what diseases you have and then sell you the drugs to treat them.
In your new book, you describe how DTC genetic testing companies have capitalized on our innate human desire to connect with or ancestors and each other. I quote you: "This industry has taken that potent, spiritual, all-too-human need to belong... and monetized it in a particularly exploitative way." But others argue that DTC genetic testing companies are merely providing a service in exchange for fair-market compensation. So where does exploitation come into the picture?
Yes, the industry provides a fee for service, but that's only part of the story. The rest of the story reveals a pernicious industry that hides its business model behind the larger science project of health and heredity. All of the major testing companies play on the idea of "lack," that we can't know who we are unless we buy information about ourselves. When you really think about it, "Who do you think you are?" is a pernicious question that suggests that we don't or can't know who we or to whom we are related without advanced data searches and testing. This existential question used to be a philosophical question; now the answers are provided by databases that acquire more valuable information than they provide in the exchange.
"It's a brilliant business model that exploits consumer naiveté."
As you've said before, consumers are actually paying to be the product because the companies are likely to profit more from selling their genetic data. Could you elaborate?
The largest databases, AncestryDNA and 23andMe, have signed lucrative agreements with biotech companies that pay them for the de-identified data of their customers. What's so valuable is the DNA combined with the family relationships. Consumers provide the family relationships and the companies link and extrapolate the results to larger and larger family trees. Combined with the genetic markers for certain diseases, or increased susceptibility to certain diseases, these databases are very valuable for biotech research.
None of that value will ever be returned to consumers except in the form of for-profit drugs. Ancestry, in particular, has removed all information about its "research partners" from its website, making it very difficult to see how it is profiting from its third-party sales. 23andMe is more open about its "two-sided business model," but encourages consumers to donate their information to science. It's a brilliant business model that exploits consumer naiveté.
A WIRED journalist wrote that "23andMe has been sharing insights gleaned from consented customer data with GSK and at least six other pharmaceutical and biotechnology firms for the past three and a half years." Is this a consumer privacy risk?
I don't see that 23andMe did anything to which consumers didn't consent, albeit through arguably unreadable terms and conditions. The part that worries me more is the 300 phenotype data points that the company has collected on its consumers through longitudinal surveys designed, as Anne Wojcicki, CEO and Co-founder of 23andMe, put it, "to circumvent medical records and just self-report."
Everyone is focused on the DNA, but it's the combination of genetic samples, genealogical information and health records that is the most potent dataset, and 23andMe has figured out a way to extract all three from consumers.


