New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
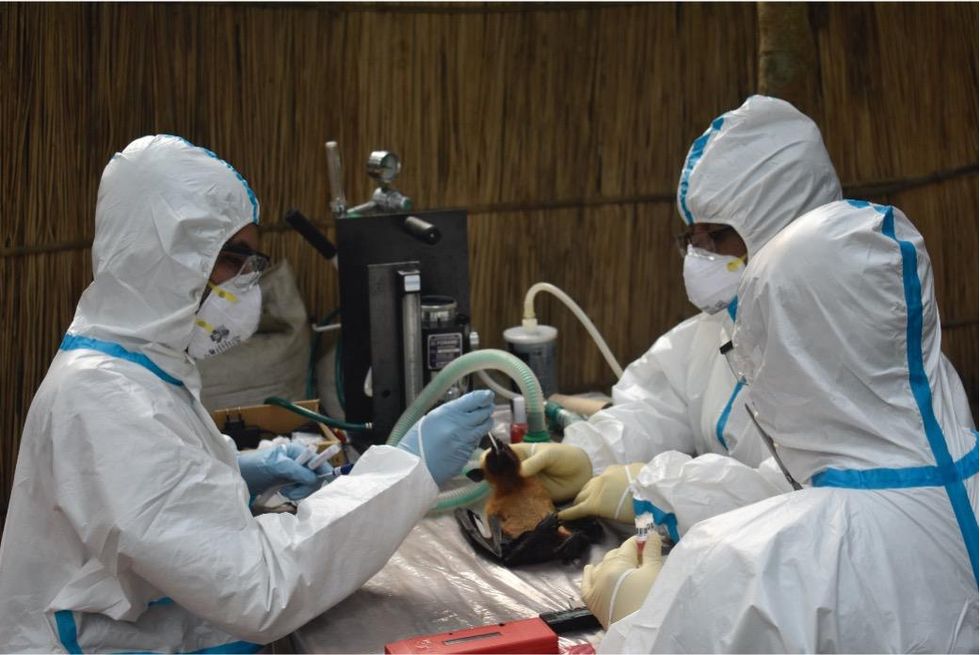
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
Scientists Just Started Testing a New Class of Drugs to Slow--and Even Reverse--Aging
Eliminating "zombie-like" cells, called senescent cells, may hold the key to slowing aging and its chronic diseases.
Imagine reversing the processes of aging. It's an age-old quest, and now a study from the Mayo Clinic may be the first ray of light in the dawn of that new era.
The immune system can handle a certain amount of senescence, but that capacity declines with age.
The small preliminary report, just nine patients, primarily looked at the safety and tolerability of the compounds used. But it also showed that a new class of small molecules called senolytics, which has proven to reverse markers of aging in animal studies, can work in humans.
Aging is a relentless assault of chronic diseases including Alzheimer's, cardiovascular disease, diabetes, and frailty. Developing one chronic condition strongly predicts the rapid onset of another. They pile on top of each other and impede the body's ability to respond to the next challenge.
"Potentially, by targeting fundamental aging processes, it may be possible to delay or prevent or alleviate multiple age-related conditions and many diseases as a group, instead of one at a time," says James Kirkland, the Mayo Clinic physician who led the study and is a top researcher in the growing field of geroscience, the biology of aging.
Getting Rid of "Zombie" Cells
One element common to many of the diseases is senescence, a kind of limbo or zombie-like state where cells no longer divide or perform many regular functions, but they don't die. Senescence is thought to be beneficial in that it inhibits the cancerous proliferation of cells. But in aging, the senescent cells still produce molecules that create inflammation both locally and throughout the body. It is a cycle that feeds upon itself, slowly ratcheting down normal body function and health.
Disease and harmful stimuli like radiation to treat cancer can also generate senescence, which is why young cancer patients seem to experience earlier and more rapid aging. The immune system can handle a certain amount of senescence, but that capacity declines with age. There also appears to be a threshold effect, a tipping point where senescence becomes a dominant factor in aging.
Kirkland's team used an artificial intelligence approach called machine learning to look for cell signaling networks that keep senescent cells from dying. To date, researchers have identified at least eight such signaling networks, some of which seem to be unique to a particular type of cell or tissue, but others are shared or overlap.
Then a computer search identified molecules known to disrupt these signaling pathways "and allow cells that are fully senescent to kill themselves," he explains. The process is a bit like looking for the right weapons in a video game to wipe out lingering zombie cells. But instead of swords, guns, and grenades, the list of biological tools so far includes experimental molecules, approved drugs, and natural supplements.
Treatment
"We found early on that targeting single components of those networks will only kill a very small minority of senescent cells or senescent cell types," says Kirkland. "So instead of going after one drug-one target-one disease, we're going after networks with combinations of drugs or drugs that have multiple targets. And we're going after every age-related disease."
The FDA is grappling with guidance for researchers wanting to conduct clinical trials on something as broad as aging rather than a single disease.
The large number of potential senolytic (i.e. zombie-neutralizing) compounds they identified allowed Kirkland to be choosy, "purposefully selecting drugs where the side effects profile was good...and with short elimination half-lives." The hit and run approach meant they didn't have to worry about maintaining a steady state of drugs in the body for an extended period of time. Some of the compounds they selected need only a half hour exposure to trigger the dying process in senescent cells, which can then take several days.
Work in mice has already shown impressive results in reversing diabetes, weight gain, Alzheimer's, cardiovascular disease and other conditions using senolytic agents.
That led to Kirkland's pilot study in humans with diabetes-related kidney disease using a three-day regimen of dasatinib, a kinase inhibitor first approved in 2006 to treat some forms of blood cancer, and quercetin, a flavonoid found in many plants and sold as a food supplement.
The combination was safe and well tolerated; it reduced the number of senescent cells in the belly fat of patients and restored their normal function, according to results published in September in the journal EBioMedicine. This preliminary paper was based on 9 patients in an ongoing study of 30 patients.
Kirkland cautions that these are initial and incomplete findings looking primarily at safety issues, not effectiveness. There is still much to be learned about the use of senolytics, starting with proof that they actually provide clinical benefit, and against what chronic conditions. The drug combinations, doses, duration, and frequency, not to mention potential risks all must be worked out. Additional studies of other diseases are being developed.
What's Next
Ron Kohanski, a senior administrator at the NIH National Institute on Aging (NIA), says the field of senolytics is so new that there isn't even a consensus on how to identify a senescent cell, and the FDA is grappling with guidance for researchers wanting to conduct clinical trials on something as broad as aging rather than a single disease.
Intellectual property concerns may temper the pharmaceutical industry's interest in developing senolytics to treat chronic diseases of aging. It looks like many mix-and-match combinations are possible, and many of the potential molecules identified so far are found in nature or are drugs whose patents have or will soon expire. So the ability to set high prices for such future drugs, and hence the willingness to spend money on expensive clinical trials, may be limited.
Still, Kohanski believes the field can move forward quickly because it often will include products that are already widely used and have a known safety profile. And approaches like Kirkland's hit and run strategy will minimize potential exposure and risk.
He says the NIA is going to support a number of clinical trials using these new approaches. Pharmaceutical companies may feel that they can develop a unique part of a senolytic combination regimen that will justify their investment. And if they don't, countries with socialized medicine may take the lead in supporting such research with the goal of reducing the costs of treating aging patients.
A New Test Aims to Objectively Measure Pain. It Could Help Legitimate Sufferers Access the Meds They Need.
Sickle cell patient Bridgett Willkie found herself being labeled an addict when she sought an opioid prescription to control her pain.
"That throbbing you feel for the first minute after a door slams on your finger."
This is how Central Florida resident Bridgett Willkie describes the attacks of pain caused by her sickle cell anemia – a genetic blood disorder in which a patient's red blood cells become shaped like sickles and get stuck in blood vessels, thereby obstructing the flow of blood and oxygen.
"I found myself being labeled as an addict and I never was."
Willkie's lifelong battle with the condition has led to avascular necrosis in both of her shoulders, hips, knees and ankles. This means that her bone tissue is dying due to insufficient blood supply (sickle cell anemia is among the medical conditions that can decrease blood flow to one's bones).
"That adds to the pain significantly," she says. "Every time my heart beats, it hurts. And the pain moves. It follows the path of circulation. I liken it to a traffic jam in my veins."
For more than a decade, she received prescriptions for Oxycontin. Then, four years ago, her hematologist – who had been her doctor for 18 years – suffered a fatal heart attack. She says her longtime doctor's replacement lacked experience treating sickle cell patients and was uncomfortable writing her a prescription for opioids. What's more, this new doctor wanted to place her in a drug rehab facility.
"Because I refused to go, he stopped writing my scripts," she says. The ensuing three months were spent at home, detoxing. She describes the pain as unbearable. "Sometimes I just wanted to die."
One of the effects of the opioid epidemic is that many legitimate pain patients have seen their opioids significantly reduced or downright discontinued because of their doctors' fears of over-prescribing addictive medications.
"I found myself being labeled as an addict and I never was...Being treated like a drug-seeking patient is degrading and humiliating," says Willkie, who adds that when she is at the hospital, "it's exhausting arguing with the doctors...You dread them making their rounds because every day they come in talking about weaning you off your meds."
Situations such as these are fraught with tension between patients and doctors, who must remain wary about the risk of over-prescribing powerful and addictive medications. Adding to the complexity is that it can be very difficult to reliably assess a patient's level of physical pain.
However, this difficulty may soon decline, as Indiana University School of Medicine researchers, led by Dr. Alexander B. Niculescu, have reportedly devised a way to objectively assess physical pain by analyzing biomarkers in a patient's blood sample. The results of a study involving more than 300 participants were published earlier this year in the journal Molecular Psychiatry.
Niculescu – who is both a professor of psychiatry and medical neuroscience at the IU School of Medicine – explains that, when someone is in severe physical pain, a blood sample will show biomarkers related to intracellular adhesion and cell-signaling mechanisms. He adds that some of these biomarkers "have prior convergent evidence from animal or human studies for involvement in pain."
Aside from reliably measuring pain severity, Niculescu says blood biomarkers can measure the degree of one's response to treatment and also assess the risk of future recurrences of pain. He believes this new method's greatest benefit, however, might be the ability to identify a number of non-opioid medications that a particular patient is likely to respond to, based on his or her biomarker profile.
Clearly, such a method could be a gamechanger for pain patients and the professionals who treat them. As of yet, health workers have been forced to make crucial decisions based on their clinical impressions of patients; such impressions are invariably subjective. A method that enables people to prove the extent of their pain could remove the stigma that many legitimate pain patients face when seeking to obtain their needed medicine. It would also improve their chances of receiving sufficient treatment.
Niculescu says it's "theoretically possible" that there are some conditions which, despite being severe, might not reveal themselves through his testing method. But he also says that, "even if the same molecular markers that are involved in the pain process are not reflected in the blood, there are other indirect markers that should reflect the distress."
Niculescu expects his testing method will be available to the medical community at large within one to three years.
Willkie says she would welcome a reliable pain assessment method. Well-aware that she is not alone in her plight, she has more than 500 Facebook friends with sickle cell disease, and she says that "all of their opioid meds have been restricted or cut" as a result of the opioid crisis. Some now feel compelled to find their opioids "on the streets." She says she personally has never obtained opioids this way. Instead, she relies on marijuana to mitigate her pain.
Niculescu expects his testing method will be available to the medical community at large within one to three years: "It takes a while for things to translate from a lab setting to a commercial testing arena."
In the meantime, for Willkie and other patients, "we have to convince doctors and nurses that we're in pain."