New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
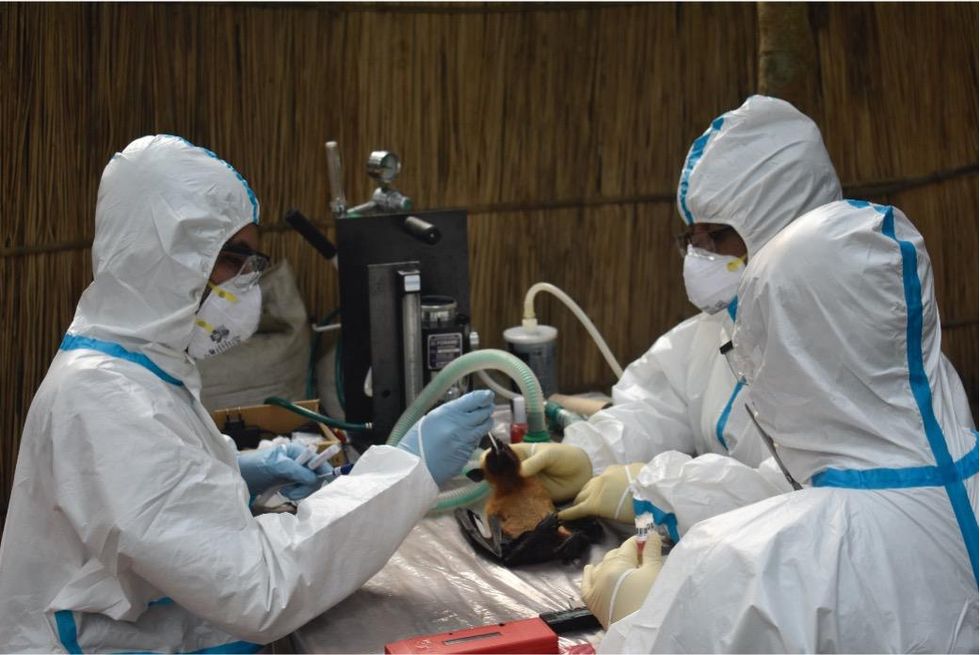
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
An astronaut peers through a portal in outer space.
What if people could just survive on sunlight like plants?
The admittedly outlandish question occurred to me after reading about how climate change will exacerbate drought, flooding, and worldwide food shortages. Many of these problems could be eliminated if human photosynthesis were possible. Had anyone ever tried it?
Extreme space travel exists at an ethically unique spot that makes human experimentation much more palatable.
I emailed Sidney Pierce, professor emeritus in the Department of Integrative Biology at the University of South Florida, who studies a type of sea slug, Elysia chlorotica, that eats photosynthetic algae, incorporating the algae's key cell structure into itself. It's still a mystery how exactly a slug can operate the part of the cell that converts sunlight into energy, which requires proteins made by genes to function, but the upshot is that the slugs can (and do) live on sunlight in-between feedings.
Pierce says he gets questions about human photosynthesis a couple of times a year, but it almost certainly wouldn't be worth it to try to develop the process in a human. "A high-metabolic rate, large animal like a human could probably not survive on photosynthesis," he wrote to me in an email. "The main reason is a lack of surface area. They would either have to grow leaves or pull a trailer covered with them."
In short: Plants have already exploited the best tricks for subsisting on photosynthesis, and unless we want to look and act like plants, we won't have much success ourselves. Not that it stopped Pierce from trying to develop human photosynthesis technology anyway: "I even tried to sell it to the Navy back in the day," he told me. "Imagine photosynthetic SEALS."
It turns out, however, that while no one is actively trying to create photosynthetic humans, scientists are considering the ways humans might need to change to adapt to future environments, either here on the rapidly changing Earth or on another planet. Rice University biologist Scott Solomon has written an entire book, Future Humans, in which he explores the environmental pressures that are likely to influence human evolution from this point forward. On Earth, Solomon says, infectious disease will remain a major driver of change. As for Mars, the big two are lower gravity and radiation, the latter of which bombards the Martian surface constantly because the planet has no magnetosphere.
Although he considers this example "pretty out there," Solomon says one possible solution to Mars' magnetic assault could leave humans not photosynthetic green, but orange, thanks to pigments called carotenoids that are responsible for the bright hues of pumpkins and carrots.
"Carotenoids protect against radiation," he says. "Usually only plants and microbes can produce carotenoids, but there's at least one kind of insect, a particular type of aphid, that somehow acquired the gene for making carotenoids from a fungus. We don't exactly know how that happened, but now they're orange... I view that as an example of, hey, maybe humans on Mars will evolve new kinds of pigmentation that will protect us from the radiation there."
We could wait for an orange human-producing genetic variation to occur naturally, or with new gene editing techniques such as CRISPR-Cas9, we could just directly give astronauts genetic advantages such as carotenoid-producing skin. This may not be as far-off as it sounds: Extreme space travel exists at an ethically unique spot that makes human experimentation much more palatable. If an astronaut already plans to subject herself to the enormous experiment of traveling to, and maybe living out her days on, a dangerous and faraway planet, do we have any obligation to provide all the protection we can?
Probably the most vocal person trying to figure out what genetic protections might help astronauts is Cornell geneticist Chris Mason. His lab has outlined a 10-phase, 500-year plan for human survival, starting with the comparatively modest goal of establishing which human genes are not amenable to change and should be marked with a "Do not disturb" sign.
To be clear, Mason is not actually modifying human beings. Instead, his lab has studied genes in radiation-resistant bacteria, such as the Deinococcus genus. They've expressed proteins called DSUP from tardigrades, tiny water bears that can survive in space, in human cells. They've looked into p53, a gene that is overexpressed in elephants and seems to protect them from cancer. They also developed a protocol to work on the NASA twin study comparing astronauts Scott Kelly, who spent a year aboard the International Space Station, and his brother Mark, who did not, to find out what effects space tends to have on genes in the first place.
In a talk he gave in December, Mason reported that 8.7 percent of Scott Kelly's genes—mostly those associated with immune function, DNA repair, and bone formation—did not return to normal after the astronaut had been home for six months. "Some of these space genes, we could engineer them, activate them, have them be hyperactive when you go to space," he said in that same talk. "When we think about having the hubris to go to a faraway planet...it seems like an almost impossible idea….but I really like people and I want us to survive for a long time, and this is the first step on the stairwell to survive out of the solar system."
What is the most important ability we could give our future selves through science?
There are others performing studies to figure out what capabilities we might bestow on the future-proof superhuman, but none of them are quite as extreme as photosynthesis (although all of them are useful). At Harvard, geneticist George Church wants to engineer cells to be resistant to viruses, such as the common cold and HIV. At Columbia, synthetic biologist Harris Wang is addressing self-sufficient humans more directly—trying to spur kidney cells to produce amino acids that are normally only available from diet.
But perhaps Future Humans author Scott Solomon has the most radical idea. I asked him a version of the classic What would be your superhero power? question: What does he see as the most important ability we could give our future selves through science?
"The empathy gene," he said. "The ability to put yourself in someone else's shoes and see the world as they see it. I think it would solve a lot of our problems."
A model of a human kidney.
Science's dream of creating perfect custom organs on demand as soon as a patient needs one is still a long way off. But tiny versions are already serving as useful research tools and stepping stones toward full-fledged replacements.
Although organoids cannot yet replace kidneys, they are invaluable tools for research.
The Lowdown
Australian researchers have grown hundreds of mini human kidneys in the past few years. Known as organoids, they function much like their full-grown counterparts, minus a few features due to a lack of blood supply.
Cultivated in a petri dish, these kidneys are still a shadow of their human counterparts. They grow no larger than one-sixth of an inch in diameter; fully developed organs are up to five inches in length. They contain no more than a few dozen nephrons, the kidney's individual blood-filtering unit, whereas a fully-grown kidney has about 1 million nephrons. And the dish variety live for just a few weeks.
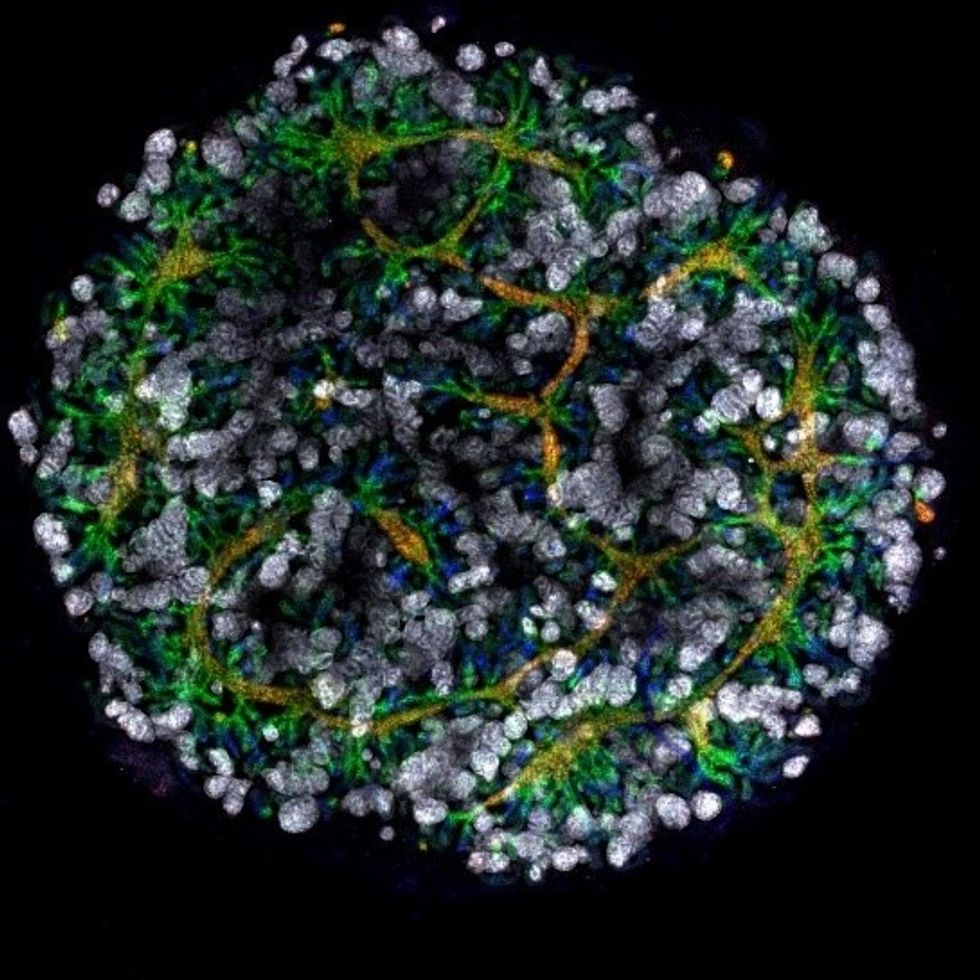
An organoid kidney created by the Murdoch Children's Institute in Melbourne, Australia.
Photo Credit: Shahnaz Khan.
But Melissa Little, head of the kidney research laboratory at the Murdoch Children's Institute in Melbourne, says these organoids are invaluable tools for research. Although renal failure is rare in children, more than half of those who suffer from such a disorder inherited it.
The mini kidneys enable scientists to better understand the progression of such disorders because they can be grown with a patient's specific genetic condition.
Mature stem cells can be extracted from a patient's blood sample and then reprogrammed to become like embryonic cells, able to turn into any type of cell in the body. It's akin to walking back the clock so that the cells regain unlimited potential for development. (The Japanese scientist who pioneered this technique was awarded the Nobel Prize in 2012.) These "induced pluripotent stem cells" can then be chemically coaxed to grow into mini kidneys that have the patient's genetic disorder.
"The (genetic) defects are quite clear in the organoids, and they can be monitored in the dish," Little says. To date, her research team has created organoids from 20 different stem cell lines.
Medication regimens can also be tested on the organoids, allowing specific tailoring for each patient. For now, such testing remains restricted to mice, but Little says it eventually will be done on human organoids so that the results can more accurately reflect how a given patient will respond to particular drugs.
Next Steps
Although these organoids cannot yet replace kidneys, Little says they may plug a huge gap in renal care by assisting in developing new treatments for chronic conditions. Currently, most patients with a serious kidney disorder see their options narrow to dialysis or organ transplantation. The former not only requires multiple sessions a week, but takes a huge toll on patient health.
Ten percent of older patients on dialysis die every year in the U.S. Aside from the physical trauma of organ transplantation, finding a suitable donor outside of a family member can be difficult.
"This is just another great example of the potential of pluripotent stem cells."
Meanwhile, the ongoing creation of organoids is supplying Little and her colleagues with enough information to create larger and more functional organs in the future. According to Little, researchers in the Netherlands, for example, have found that implanting organoids in mice leads to the creation of vascular growth, a potential pathway toward creating bigger and better kidneys.
And while Little acknowledges that creating a fully-formed custom organ is the ultimate goal, the mini organs are an important bridge step.
"This is just another great example of the potential of pluripotent stem cells, and I am just passionate to see it do some good."