New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
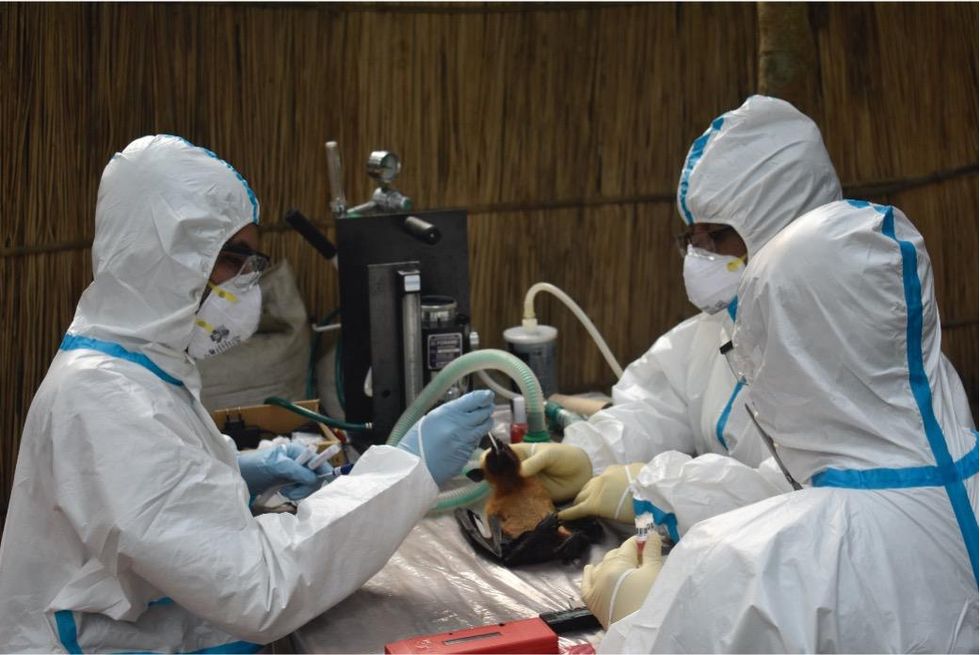
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
Couples Facing Fertility Treatments Should Beware of This
Couples facing infertility should be savvy about the add-on procedures offered by some fertility clinics.
When Jane Stein and her husband used in-vitro fertilization in 2001 to become pregnant with twins, her fertility clinic recommended using a supplemental procedure called intracytoplasmic sperm injection (ICSI), known in fertility lingo as "ix-see."
'Add-on' fertility procedures are increasingly coming under scrutiny for having a high cost and low efficacy rate.
During IVF, an egg and sperm are placed in a petri dish together with the hope that a sperm will seek out and fertilize the egg. With ICSI, doctors inject sperm directly into the egg.
Stein, whose name has been changed to protect her privacy, agreed to try it. Her twins are now 16, but while 17 years have gone by since that procedure, the efficacy of ICSI is still unclear. In other words, while Stein succeeded in having children, it may not have been because of ICSI. It may simply have been because she did IVF.
The American Society for Reproductive Medicine has concluded, "There are no data to support the routine use of ICSI for non-male factor infertility." That is, ICSI can help couples have a baby when the issue is male infertility. But when it's not, the evidence of its effectiveness is lacking. And yet the procedure is being used more and more, even when male infertility is not the issue. Some 40 percent of fertility treatments in Europe, Asia and the Middle East now use ICSI, according to a world report released in 2016 by the International Committee for Monitoring Assisted Reproductive Technologies. In the Middle East, the figure is actually 100 percent, the report said.
ICSI is just one of many supplemental procedures, or 'add-ons,' increasingly coming under scrutiny for having a high cost and low efficacy rate. They cost anywhere from a couple of hundred dollars to several thousand – ICSI costs $2,000 to $3,000 -- hiking up the price of what is already a very costly endeavor. And many don't even work. Worse, some actually cause harm.
It's no surprise couples use them. They promise to increase the chance of conceiving. For patients who desperately want a child, money is no object. The Human Fertilization and Embryology Authority (HFEA) in the U.K. found that some 74 percent of patients who received fertility treatments over the last two years were given at least one type of add-on. Now, fertility associations in the U.S. and abroad have begun issuing guidance about which add-ons are worth the extra cost and which are not.
"Many IVF add-ons have little in the way of conclusive evidence supporting their role in successful IVF treatment," said Professor Geeta Nargund, medical director of CREATE Fertility and Lead Consultant for reproductive medicine at St George's Hospital, London.
The HFEA has actually rated these add-ons, indicating which procedures are effective and safe. Some treatments were rated 'red' because they were considered to have insufficient evidence to justify their use. These include assisted hatching, which uses acid or lasers to make a hole in the surrounding layer of proteins to help the embryo hatch; intrauterine culture, where a device is inserted into the womb to collect and incubate the embryo; and reproductive immunology, which suppresses the body's natural immunity so that it accepts the embryo.
"Fertility care is a highly competitive market. In a private system, offering add-ons may discern you from your neighboring clinic."
For some treatments, the HFEA found there is evidence that they don't just fail to work, but can even be harmful. These procedures include ICSI used when male infertility is not at issue, as well as a procedure called endometrial scratching, where the uterus is scratched, not unlike what would happen with a biopsy, to stimulate the local uterine immune system.
And then for some treatments, there is conflicting evidence, warranting further research. These include artificial egg activation by calcium ionophore, elective freezing in all cycles, embryo glue, time-lapse imaging and pre-implantation genetic testing for abnormal chromosomes on day 5.
"Currently, there is very little evidence to suggest that many of the add-ons could increase success rates," Nargund said. "Indeed, the HFEA's assessment of add-on treatments concluded that none of the add-ons could be given a 'green' rating, due to a lack of conclusive supporting research."
So why do fertility clinics offer them?
"Fertility care is a highly competitive market," said Professor Hans Evers, editor-in-chief of the journal Human Reproduction. "In a private system, offering add-ons may discern you from your neighboring clinic. The more competition, the more add-ons. Hopefully the more reputable institutions will only offer add-ons (for free) in the context of a randomized clinical trial."
The only way for infertile couples to know which work and which don't is the guidance released by professional organizations like the ASRM, and through government regulation in countries that have a public health care system.
The problem is, infertile couples will sometimes do anything to achieve a pregnancy.
"They will stand on their heads if this is advocated as helpful. Someone has to protect them," Evers said.
In the Netherlands, where Evers is based, the national health care system tries to make the best use of the limited resources it has, so it makes sure the procedures it's funding actually work, Evers said.
"We have calculated that to serve a population of 17 million, we need 13 IVF clinics, and we have 13," he said. "We as professionals discuss and try to agree on the value of newly proposed add-ons, and we will implement only those that are proven effective and safe."
Likewise, in the U.K., there's been a lot of squawking about speculative add-ons because the government, or National Health Service, pays for them. In the U.S., it's private insurers or patients' own cash.
"The [U.K.] government takes a very close look at what therapies they are offering and what the evidence is around offering the therapy," said Alan Penzias, who chairs the Practice Committee of the ASRM. It wants to make sure the treatments it is funding are at least worth the money.
ICSI is a case in point. Originally intended for male infertility, it's now being applied across the board because fertility clinics didn't want couples to pay $10,000 to $15,000 and wind up with no embryos.
"It is so disastrous to have no fertilization whatsoever, clinics started to make this bargain with their patients, saying, 'Well, listen, even though it's not indicated, what we would like to do is to take half of your eggs and do the ICSI procedure, and half we'll do conventional insemination just to make sure,'" he said. "It's a disaster if you have no embryos, and now you're out 10 to 12 thousand dollars, so for a small added fee, we can do the injection just to guard against that."
In the Netherlands, the national health care system tries to make the best use of its limited resources, so it makes sure the procedures it's funding actually work.
Clinics offer it where they see lower rates of fertilization, such as with older women or in cases where induced ovulation results in just two or three eggs instead of, say, 13. Unfortunately, ICSI may result in a higher fertilization rate, but it doesn't result in a higher live birth rate, according to a study last year in Human Reproduction, so couples wind up paying for a procedure that doesn't even result in a child.
Private insurers in the U.S. are keen to it. Penzia, who is also an associate professor of obstetrics, gynecology and reproductive biology at Harvard Medical School and works as a reproductive endocrinology and infertility specialist at Boston IVF, said Massachusetts requires that insurers cover infertility treatments. But when he submits claims for ICSI, for instance, insurers now want to see two sperm counts and proof that the man has seen a urologist.
"They want to make sure we're doing it for male factor (infertility)," he said. "That's not unreasonable, because the insurance company is taking the burden of this."
Epigenetic therapeutics could revolutionize medicine in the coming decades. (© kentoh/Adobe)
How exactly does your DNA make you who you are?
It's because of epigenetics that identical twins can actually look different and develop different diseases.
Just as software developers don't write apps out of ones and zeros, the interesting parts of the human genome aren't written merely in As, Ts, Cs and Gs. Yes, these are the fundamental letters that make up our DNA and encode the proteins that make our cells function, but the story doesn't end there.
Our cells possess amazing abilities, like eating invading bacteria or patching over a wound, and these abilities require the coordinated action of hundreds, if not thousands, of proteins. Epigenetics, the study of gene expression, examines how multiple genes work at once to make these biological processes happen.
It's because of epigenetics that identical twins – who possess identical DNA -- can actually look different and develop different diseases. Their environments may influence the expression of their genes in unique ways. For example, a research study in mice found that maternal exposure to a chemical called bisphenol A (BPA) resulted in drastic differences between genetically identical offspring. BPA exposure increased the likelihood that a certain gene was turned on, which led to the birth of yellow mice who were prone to obesity. Their genetically identical siblings who were not exposed to BPA were thinner and born with brown fur.

These three mice are genetically identical. Epigenetic differences, however, result in vastly different phenotypes.
(© 1994 Nature Publishing Group, Duhl, D.)
This famous mouse experiment is just one example of how epigenetics may transform medicine in the coming years. By studying the way genes are turned on and off, and maybe even making those changes ourselves, scientists are beginning to approach diseases like cancer in a completely new way.
With few exceptions, most of the 1 trillion cells that make up your body contain the same DNA instructions as all the others. How does each cell in your body know what it is and what it has to do? One of the answers appears to lie in epigenetic regulation. Just as everyone at a company may have access to all the same files on the office Dropbox, the accountants will put different files on their desktop than the lawyers do.
Our cells prioritize DNA sequences in the same way, even storing entire chromosomes that aren't needed along the wall of the nucleus, while keeping important pieces of DNA in the center, where it is most accessible to be read and used. One of the ways our cells prioritize certain DNA sequences is through methylation, a process that inactivates large regions of genes without editing the underlying "file" itself.
As we learn more about epigenetics, we gain more opportunities to develop therapeutics for a broad range of human conditions, from cancer to metabolic disorders. Though there have not been any clinical applications of epigenetics to immune or metabolic diseases yet, cancer is one of the leading areas, with promising initial successes.
One of the challenges of cancer treatments is that different patients may respond positively or negatively to the same treatment. With knowledge of epigenetics, however, doctors could conduct diagnostic tests to identify a patient's specific epigenetic profile and determine the best treatment for him or her. Already, commercial kits are available that help doctors screen glioma patients for an epigenetic biomarker called MGMT, because patients with this biomarker have shown high rates of success with certain kinds of treatments.
Other epigenetic advances go beyond personalized screening to treatments targeting the mechanism of disease. Some epigenetic drugs turn on genes that help suppress tumors, while others turn on genes that reveal the identity of tumor cells to the immune system, allowing it to attack cancerous cells.
Direct, targeted control of your epigenome could allow doctors to reprogram cancerous or aging cells.
The study of epigenetics has also been fundamental to the field of aging research. The older you get, the more methylation marks your DNA carries, and this has led to the distinction between biological aging, or the state of your cells, and chronological aging, or how old you actually are.
Just as our DNA can get miscopied and accumulate mutations, errors in DNA methylation can lead to so-called "epimutations". One of the big hypotheses in aging research today is that the accumulation of these random epimutations over time is responsible for what we perceive as aging.
Studies thus far have been correlative - looking at several hundred sites of epigenetic modifications in a person's cell, scientists can now roughly discern the age of that person. The next set of advances in the field will come from learning what these epigenetic changes individually do by themselves, and if certain methylations are correlated with cellular aging. General diagnostic terms like "aging" could be replaced with "abnormal methylation at these specific locations," which would also open the door to new therapeutic targets.
Direct, targeted control of your epigenome could allow doctors to reprogram cancerous or aging cells. While this type of genetic surgery is not feasible just yet, current research is bringing that possibility closer. The Cas9 protein of genome-editing CRISPR/Cas9 fame has been fused with epigenome modifying enzymes to target epigenetic modifications to specific DNA sequences.
A therapeutic of this type could theoretically undo a harmful DNA methylation, but would also be competing with the cell's native machinery responsible for controlling this process. One potential approach around this problem involves making beneficial synthetic changes to the epigenome that our cells do not have the capacity to undo.
Also fueling this frontier is a new approach to understanding disease itself. Scientists and doctors are now moving beyond the "one defective gene = one disease" paradigm. Because lots of diseases are caused by multiple genes going haywire, epigenetic therapies could hold the key to new types of treatments by targeting multiple defective genes at once.
Scientists are still discovering which epigenetic modifications are responsible for particular diseases, and engineers are building new tools for epigenome editing. Given the proliferation of work in these fields within the last 10 years, we may see epigenetic therapeutics emerging within the next couple of decades.

