New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
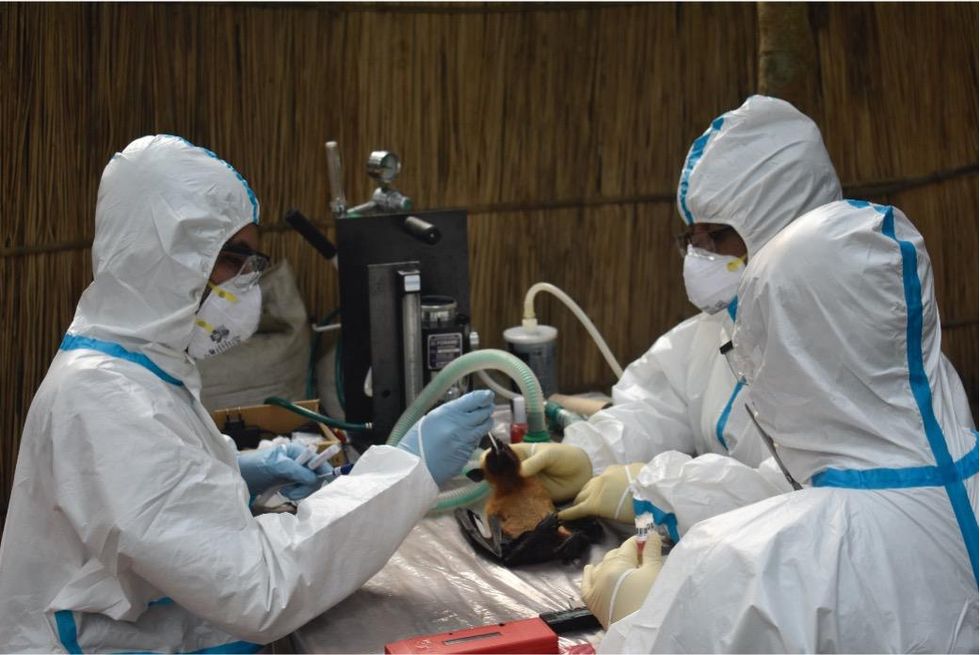
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
This Brain Doc Has a “Repulsive” Idea to Make Football Safer
A football player making a tackle during a game. (© Joe/Fotolia)
What do football superstars Tom Brady, Drew Brees, Philip Rivers, and Adrian Peterson all have in common? Last year they wore helmets that provided the poorest protection against concussions in all the NFL.
"You're only as protected as well as the worst helmet that's out there."
A Dangerous Policy
Football helmets are rated on a one-star to five-star system based on how well they do the job of protecting the player. The league has allowed players to use their favorites, regardless of the star rating.
The Oxford-trained neuroscientist Ray Colello conducted a serious analysis of just how much the protection can vary between each level of star rating. Colello and his team of graduate students sifted through two seasons of game video to identify which players were wearing what helmets. There was "a really good correlation with position, but the correlation is much more significant based on age."
"The average player in the NFL is 26.6 years old, but the average age of a player wearing a one-star helmet is 34. And for anyone who knows football, that's ancient," the brain doc says. "Then for our two-star helmet, it's 32; and for a three-star helmet it's 29." Players were sticking with the helmets they were familiar with in college, despite the fact that equipment had improved considerably in recent years.
"You're only as protected as well as the worst helmet that's out there," Colello explains. Offering an auto analogy, he says, "It's like, if you run into the back of a Pinto, even if you are in a five-star Mercedes, that gas tank may still explode and you are still going to die."
It's one thing for a player to take a risk at scrambling his own brain; it's another matter to put a teammate or opponent at needless risk. Colello published his analysis early last year and the NFL moved quickly to ban the worst performing helmets, starting next season.
Some of the 14 players using the soon-to-be-banned helmets, like Drew Brees and Philip Rivers, made the switch to a five-star helmet at the start of training camp and stayed with it. Adrian Peterson wore a one-star helmet throughout the season.
Tom Brady tried but just couldn't get comfortable with a new bonnet and, after losing a few games, switched back to his old one in the middle of the season; he says he's going to ask the league to "grandfather in" his old helmet so he can continue to use it.
As for Colello, he's only just getting started. The brain doc has a much bigger vision for the future of football safety. He wants to prevent concussions from even occurring in the first place by creating an innovative new helmet that's unlike anything the league has ever seen.

Oxford-trained neuroscientist Ray Colello is on a mission to make football safer.
(Photo credit: VCU public affairs)
"A Force Field" of Protection
His inspiration was serendipitous; he was at home watching a football game on TV when Denver Bronco's receiver Wes Welker was hit, lay flat on the field with a concussion, and was carted off. As a commercial flickered on the screen, he ambled into the kitchen for another beer. "What those guys need is a force field protecting them," he thought to himself.
Like so many households, the refrigerator door was festooned with magnets holding his kids' school work in place. And in that eureka moment the idea popped into his head: "Maybe the repulsive force of magnets can put a break on an impact before it even occurs." Colello has spent the last few years trying to turn his concept into reality.
Newton's laws of physics – mass and speed – play out graphically in a concussion. The sudden stop of a helmet-to-helmet collision can shake the brain back and forth inside the skull like beans in a maraca. Dried beans stand up to the impact, making their distinctive musical sound; living brain tissue is much softer and not nearly so percussive. The resulting damage is a concussion.
The risk of that occurring is greater than you might think. Researchers using accelerometers inside helmets have determined that a typical college football player experiences about 600 helmet-to-helmet contacts during a season of practice and games. Each hit generates a split second peak g-force of 20 to 150 within the helmet and the odds of one causing a concussion increase sharply over 100 gs of force.
By comparison, astronauts typically experience a maximum sustained 3gs during lift off and most humans will black out around 9gs, which is why fighter pilots wear special pressure suits to counter the effects.
"It stretches the time line of impact quite dramatically. In fact in most instances, it doesn't even hit."
The NFL's fastest player, Chris Johnson, can run 19.3 mph. A collision at that speed "produces 120gs worth of force," Colello explains. "But if you can extend that time of impact by just 5 milliseconds (from 12 to 17msec) you'll shift that g-force down to 84. There is a very good chance that he won't suffer a concussion."
The neuroscientist dived into learning all he could about the physics magnets. It turns out that the most powerful commercially available magnet is an alloy made of neodymium, iron, and boron. The elements can be mixed and glued together in any shape and then an electric current is run through to make it magnetic; the direction of the current establishes the north-south poles.
A 1-pound neodymium magnet can repulse 600 times its own weight, even though the magnetic field extends less than an inch. That means it can push back a magnet inside another helmet but not affect the brain.
Crash Testing the Magnets
Colello couldn't wait to see if his idea panned out. With blessing from his wife to use their credit card, he purchased some neodymium magnets and jury-rigged experiments at home.
The reinforced plastics used in football helmets don't affect the magnetic field. And the small magnets stopped weights on gym equipment that were dropped from various heights. "It stretches the time line of impact quite dramatically. In fact in most instances, it doesn't even hit," says Colello. "We are dramatically shifting the curve" of impact.
Virginia Commonwealth University stepped in with a $50,000 innovation grant to support the next research steps. The professor ordered magnets custom-designed to fit the curvature of space inside the front and sides of existing football helmets. That makes it impossible to install them the wrong way, and ensures the magnets' poles will always repel and not attract. It adds about a pound and a half to the weight of the helmet.

a) The brain in a helmet. b) Placing the magnet. c) Measuring the impact of a helmet-to-helmet collision. d) How magnets reduce the force of impact.
(Courtesy Ray Colello)
Colello rented crash test dummy heads crammed with accelerometers and found that the magnets performed equally well at slowing collisions when fixed to a pendulum in a test that approximated a helmet and head hitting a similarly equipped helmet. It impressively reduced the force of contact.
The NFL was looking for outside-the-box thinking to prevent concussions. It was intrigued by Colello's approach and two years ago invited him to submit materials for review. To be fair to all entrants, the league proposed to subject all entries to the same standard crush test to see how well each performed in lessening impact. The only trouble was, Colello's approach was designed to avoid collisions, not lessen their impact. The test wouldn't have been a valid evaluation and he withdrew from consideration.
But Colello's work caught the attention of Stefan Duma, an engineering professor at Virginia Tech who developed the five-star rating system for football helmets.
"In theory it makes sense to use [the magnets] to slow down or reduce acceleration, that's logical," says Duma. He believes current helmet technology is nearing "the end of the physics barrier; you can only absorb so much energy in so much space," so the field is ripe for new approaches to improve helmet technology.
However, one of Duma's concerns is whether magnets "are feasible from a weight standpoint." Most helmets today weigh between two and four pounds, and a sufficiently powerful magnet might add too much weight. One possibility is using an electromagnet, which potentially could be lighter and more powerful, particularly if the power supply could be carried lower in the body, say in the shoulder pads.
Colello says his lab tests are promising enough that the concept needs to be tried out on the playing field. "We need to make enough helmets for two teams to play each other in a regulation-style game and measure the impact forces that are generated on each, and see if there is a significant reduction." He is waiting to hear from the National Institutes of Health on a grant proposal to take that next step toward dramatically reducing the risk of concussions in the NFL.
Just five milliseconds could do it.
Genetically Sequencing Healthy Babies Yielded Surprising Results
A newborn and mother in the hospital - first touch. (© martin81/Shutterstock)
Today in Melrose, Massachusetts, Cora Stetson is the picture of good health, a bubbly precocious 2-year-old. But Cora has two separate mutations in the gene that produces a critical enzyme called biotinidase and her body produces only 40 percent of the normal levels of that enzyme.
In the last few years, the dream of predicting and preventing diseases through genomics, starting in childhood, is finally within reach.
That's enough to pass conventional newborn (heelstick) screening, but may not be enough for normal brain development, putting baby Cora at risk for seizures and cognitive impairment. But thanks to an experimental study in which Cora's DNA was sequenced after birth, this condition was discovered and she is being treated with a safe and inexpensive vitamin supplement.
Stories like these are beginning to emerge from the BabySeq Project, the first clinical trial in the world to systematically sequence healthy newborn infants. This trial was led by my research group with funding from the National Institutes of Health. While still controversial, it is pointing the way to a future in which adults, or even newborns, can receive comprehensive genetic analysis in order to determine their risk of future disease and enable opportunities to prevent them.
Some believe that medicine is still not ready for genomic population screening, but others feel it is long overdue. After all, the sequencing of the Human Genome Project was completed in 2003, and with this milestone, it became feasible to sequence and interpret the genome of any human being. The costs have come down dramatically since then; an entire human genome can now be sequenced for about $800, although the costs of bioinformatic and medical interpretation can add another $200 to $2000 more, depending upon the number of genes interrogated and the sophistication of the interpretive effort.

Two-year-old Cora Stetson, whose DNA sequencing after birth identified a potentially dangerous genetic mutation in time for her to receive preventive treatment.
(Photo courtesy of Robert Green)
The ability to sequence the human genome yielded extraordinary benefits in scientific discovery, disease diagnosis, and targeted cancer treatment. But the ability of genomes to detect health risks in advance, to actually predict the medical future of an individual, has been mired in controversy and slow to manifest. In particular, the oft-cited vision that healthy infants could be genetically tested at birth in order to predict and prevent the diseases they would encounter, has proven to be far tougher to implement than anyone anticipated.
But in the last few years, the dream of predicting and preventing diseases through genomics, starting in childhood, is finally within reach. Why did it take so long? And what remains to be done?
Great Expectations
Part of the problem was the unrealistic expectations that had been building for years in advance of the genomic science itself. For example, the 1997 film Gattaca portrayed a near future in which the lifetime risk of disease was readily predicted the moment an infant is born. In the fanfare that accompanied the completion of the Human Genome Project, the notion of predicting and preventing future disease in an individual became a powerful meme that was used to inspire investment and public support for genomic research long before the tools were in place to make it happen.
Another part of the problem was the success of state-mandated newborn screening programs that began in the 1960's with biochemical tests of the "heel-stick" for babies with metabolic disorders. These programs have worked beautifully, costing only a few dollars per baby and saving thousands of infants from death and severe cognitive impairment. It seemed only logical that a new technology like genome sequencing would add power and promise to such programs. But instead of embracing the notion of newborn sequencing, newborn screening laboratories have thus far rejected the entire idea as too expensive, too ambiguous, and too threatening to the comfortable constituency that they had built within the public health framework.
"What can you find when you look as deeply as possible into the medical genomes of healthy individuals?"
Creating the Evidence Base for Preventive Genomics
Despite a number of obstacles, there are researchers who are exploring how to achieve the original vision of genomic testing as a tool for disease prediction and prevention. For example, in our NIH-funded MedSeq Project, we were the first to ask the question: "What can you find when you look as deeply as possible into the medical genomes of healthy individuals?"
Most people do not understand that genetic information comes in four separate categories: 1) dominant mutations putting the individual at risk for rare conditions like familial forms of heart disease or cancer, (2) recessive mutations putting the individual's children at risk for rare conditions like cystic fibrosis or PKU, (3) variants across the genome that can be tallied to construct polygenic risk scores for common conditions like heart disease or type 2 diabetes, and (4) variants that can influence drug metabolism or predict drug side effects such as the muscle pain that occasionally occurs with statin use.
The technological and analytical challenges of our study were formidable, because we decided to systematically interrogate over 5000 disease-associated genes and report results in all four categories of genetic information directly to the primary care physicians for each of our volunteers. We enrolled 200 adults and found that everyone who was sequenced had medically relevant polygenic and pharmacogenomic results, over 90 percent carried recessive mutations that could have been important to reproduction, and an extraordinary 14.5 percent carried dominant mutations for rare genetic conditions.
A few years later we launched the BabySeq Project. In this study, we restricted the number of genes to include only those with child/adolescent onset that could benefit medically from early warning, and even so, we found 9.4 percent carried dominant mutations for rare conditions.
At first, our interpretation around the high proportion of apparently healthy individuals with dominant mutations for rare genetic conditions was simple – that these conditions had lower "penetrance" than anticipated; in other words, only a small proportion of those who carried the dominant mutation would get the disease. If this interpretation were to hold, then genetic risk information might be far less useful than we had hoped.
Suddenly the information available in the genome of even an apparently healthy individual is looking more robust, and the prospect of preventive genomics is looking feasible.
But then we circled back with each adult or infant in order to examine and test them for any possible features of the rare disease in question. When we did this, we were surprised to see that in over a quarter of those carrying such mutations, there were already subtle signs of the disease in question that had not even been suspected! Now our interpretation was different. We now believe that genetic risk may be responsible for subclinical disease in a much higher proportion of people than has ever been suspected!
Meanwhile, colleagues of ours have been demonstrating that detailed analysis of polygenic risk scores can identify individuals at high risk for common conditions like heart disease. So adding up the medically relevant results in any given genome, we start to see that you can learn your risks for a rare monogenic condition, a common polygenic condition, a bad effect from a drug you might take in the future, or for having a child with a devastating recessive condition. Suddenly the information available in the genome of even an apparently healthy individual is looking more robust, and the prospect of preventive genomics is looking feasible.
Preventive Genomics Arrives in Clinical Medicine
There is still considerable evidence to gather before we can recommend genomic screening for the entire population. For example, it is important to make sure that families who learn about such risks do not suffer harms or waste resources from excessive medical attention. And many doctors don't yet have guidance on how to use such information with their patients. But our research is convincing many people that preventive genomics is coming and that it will save lives.
In fact, we recently launched a Preventive Genomics Clinic at Brigham and Women's Hospital where information-seeking adults can obtain predictive genomic testing with the highest quality interpretation and medical context, and be coached over time in light of their disease risks toward a healthier outcome. Insurance doesn't yet cover such testing, so patients must pay out of pocket for now, but they can choose from a menu of genetic screening tests, all of which are more comprehensive than consumer-facing products. Genetic counseling is available but optional. So far, this service is for adults only, but sequencing for children will surely follow soon.
As the costs of sequencing and other Omics technologies continue to decline, we will see both responsible and irresponsible marketing of genetic testing, and we will need to guard against unscientific claims. But at the same time, we must be far more imaginative and fast moving in mainstream medicine than we have been to date in order to claim the emerging benefits of preventive genomics where it is now clear that suffering can be averted, and lives can be saved. The future has arrived if we are bold enough to grasp it.
Funding and Disclosures:
Dr. Green's research is supported by the National Institutes of Health, the Department of Defense and through donations to The Franca Sozzani Fund for Preventive Genomics. Dr. Green receives compensation for advising the following companies: AIA, Applied Therapeutics, Helix, Ohana, OptraHealth, Prudential, Verily and Veritas; and is co-founder and advisor to Genome Medical, Inc, a technology and services company providing genetics expertise to patients, providers, employers and care systems.

