A Million Patients Have Innovated Their Own Medical Solutions, And Doctors Are Terrified

Diabetes patient advocate Renza Scibilia with her continuous glucose monitor, used in innovative DIY health technology.
In the fall of 2017, patient advocate Renza Scibilia told a conference of endocrinologists in Australia about new, patient-developed artificial pancreas technology that helped her manage her Type 1 diabetes.
"Because it's not a regulated product, some [doctors] were worried and said 'What if it goes wrong?'"
"They were in equal measure really interested and really scared," recalled Scibilia. "Because it's not a regulated product, some were worried and said 'What if it goes wrong? What is my liability going to be?'"
That was two years ago. Asked if physicians have been more receptive to the same "looping" technology now that its benefits have been supported by considerable data (as Leapsmag pointed out in May), Scibilia said, "No. Clinicians are still really insecure. They're always going to be reluctant to accept consumer-driven technology."
This exemplifies a major challenge to the growing Do-It-Yourself (DIY) biohealth movement: physicians are unnerved and worried about innovations developed by patients and other consumers that haven't been tested in elaborate clinical trials or sanctioned by regulatory authorities.
"It's difficult for patients who develop new health technology to demonstrate the advantage in a way that physicians would accept." said Howard DeMonaco, visiting scientist at MIT's Sloan School of Management. "New approaches to the treatment of diseases are by definition suspect to clinicians. Most are risk averse unless there is a substantial advantage to the new approach and the risks in doing so appear to be minimized."
Nevertheless, the DIY biohealth movement is booming. About a million people reported that they created medical innovations to address their own medical needs in surveys conducted from 2010-2015 in the U.S., U.K., Finland, Canada and South Korea.
Add in other DIY health innovations created in homes, community biolabs and "Maker" health fairs, and it's clear that health care providers are increasingly confronted with medical devices, information technology, and even medications that were developed in unconventional settings and lack the blessing of regulatory authorities.
Researchers in Portugal have tried to spread the word about many of these solutions on the Patent Innovations website, which has more than 500 examples, ranging from a 3-D printed arm and hand to a sensor device that warns someone when an osteomy bag is full.
When Reddit asked medical professionals, "What is the craziest DIY health treatment you've seen a patient attempt?" thousands shared horror stories.
But even in this era of patient empowerment, more widespread use of DIY health solutions still depends upon the approval and cooperation of physicians, nurses and other caregivers. And health care providers still lack awareness of promising patient-developed innovations, according to Dr. Joyce Lee, a pediatric endocrinologist at the University of Michigan who advocates involving patients in the design of healthcare technology. "Most physicians are scared of what they don't know," she said.
They're also understandably worried about patients who don't know what they're doing and make irresponsible decisions. When Reddit asked medical professionals, "What is the craziest DIY health treatment you've seen a patient attempt?" thousands shared horror stories, including a man who poked a hole in his belly button with a knitting needle to relieve gas.
Yet DeMonaco and Lee think it's possible to start bridging the gaps between responsible patient innovators and skeptical doctors as well as unprepared regulatory systems.
One obstacle to consumer-driven health innovations is that clinical trials to prove their safety and effectiveness are expensive and time-consuming, as De Monaco points out in a recent article. He and his colleagues suggested that low-cost clinical trials by and for patients could help address this challenge. They urged patients to publish their own research and detail the impact of innovations on their own health, and create databases that incorporate the findings of other patients.
For example, Adam Brown, who has Type 1 diabetes, compared the effects of low and high carbohydrate diets on his blood sugar management, and conveyed the results in an online journal. "Sharing the information allowed others to copy the experiment," the article noted, suggesting that this could be a model to create multi-patient trials that could be "analyzed by expert patients and/or by professionals."
Asked how to convince health care providers to consider such research, DeMonaco cited the example of doctors prescribing "off label" drugs for purposes that aren't approved by the FDA. "The secret to off label use, like any other user innovation, is dissemination," he said. Sharing case reports and other low-cost research serves to disseminate the information "in a way that is comfortable for physicians," he said, and urged patient innovators to take the same approach.
The FDA regulates commercial products and has no authority if consumers want to use medical devices, medications, or information systems that they find on their own.
Physicians should also be encouraged to engage in patient-driven research, said Dr. Lee. She suggests forming "maker spaces in which patients and physicians are involved in designing personalized technology for chronic diseases. In my vision, patient peers would build, iterate, and learn from each other and the doctor would be part of the team, constantly assessing and evaluating the technology and facilitating the process."
Some kind of regulatory oversight of DIY health technology is also necessary, said Todd Kuiken, senior research scholar at NC State and former principal investigator at the Woodrow Wilson Center's Synthetic Biology Project.
The FDA regulates commercial products and has no authority if consumers want to use medical devices, medications, or information systems that they find on their own. But that doesn't stop regulators from worrying about patients who use them. For example, the FDA issued a warning about diabetes looping technology earlier this year after one diabetic was hospitalized with hypoglycemia.
Kuiken, for one, believes that citizen-driven innovation requires oversight "to move forward." He suggested that Internal Review Boards, with experts on medical technology, safety and ethics, could play a helpful role in validating the work of patient innovators and others engaged in DIY health research. "As people are developing health products, there would be experts available to take a look and check in," he said.
Kuiken pointed out that in native American territories, tribally based IRBs working with the national Indian Health Services help to oversee new health science research. The model could be applied more broadly.
He also offered hope to those who want to integrate the current health regulatory structure into the ecosystem of DIY health innovations. "I didn't expect people from the FDA or NIH to show up" he said about a workshop on citizen-driven biomedical research that he helped organize at the Wilson Center last year. But senior officials from both agencies attended.
He indicated they "were open to new ideas." While he wouldn't disclose contributions made by individual participants in the workshop, he said the government staffers were "very interested in figuring out how to engage with citizen health innovators, to build bridges with the DIY community."
"Why should we wait for regulatory bodies? Why wait for trials that take too long?"
Time will tell whether those bridges will be built quickly enough to increase the comfort of physicians with health innovations developed by patients and other consumers. In the meantime, DIY health innovators like patient advocate Scibilia are undeterred.
"Why should we wait for regulatory bodies?" she asked. "Why wait for trials that take too long? There are plenty of data out there indicating the [diabetes looping] technology works. So we're just going to do it. We're not waiting."
DNA- and RNA-based electronic implants may revolutionize healthcare
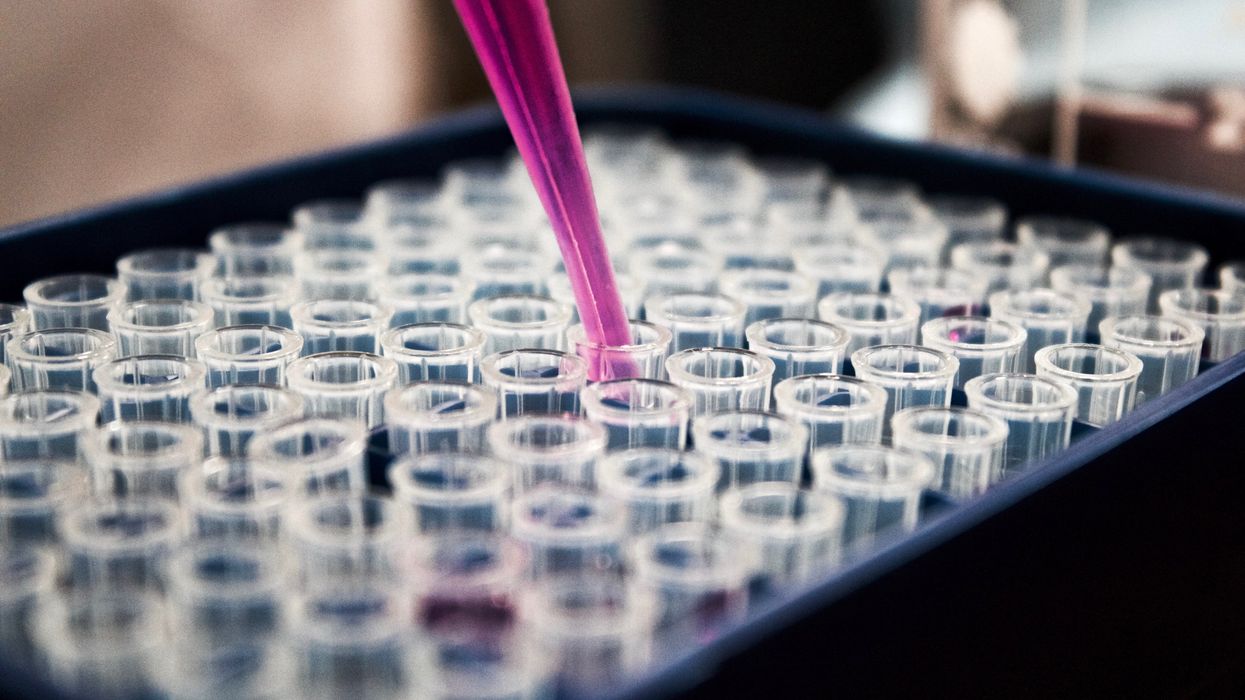
The test tubes contain tiny DNA/enzyme-based circuits, which comprise TRUMPET, a new type of electronic device, smaller than a cell.
Implantable electronic devices can significantly improve patients’ quality of life. A pacemaker can encourage the heart to beat more regularly. A neural implant, usually placed at the back of the skull, can help brain function and encourage higher neural activity. Current research on neural implants finds them helpful to patients with Parkinson’s disease, vision loss, hearing loss, and other nerve damage problems. Several of these implants, such as Elon Musk’s Neuralink, have already been approved by the FDA for human use.
Yet, pacemakers, neural implants, and other such electronic devices are not without problems. They require constant electricity, limited through batteries that need replacements. They also cause scarring. “The problem with doing this with electronics is that scar tissue forms,” explains Kate Adamala, an assistant professor of cell biology at the University of Minnesota Twin Cities. “Anytime you have something hard interacting with something soft [like muscle, skin, or tissue], the soft thing will scar. That's why there are no long-term neural implants right now.” To overcome these challenges, scientists are turning to biocomputing processes that use organic materials like DNA and RNA. Other promised benefits include “diagnostics and possibly therapeutic action, operating as nanorobots in living organisms,” writes Evgeny Katz, a professor of bioelectronics at Clarkson University, in his book DNA- And RNA-Based Computing Systems.
While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output.
Adamala’s research focuses on developing such biocomputing systems using DNA, RNA, proteins, and lipids. Using these molecules in the biocomputing systems allows the latter to be biocompatible with the human body, resulting in a natural healing process. In a recent Nature Communications study, Adamala and her team created a new biocomputing platform called TRUMPET (Transcriptional RNA Universal Multi-Purpose GatE PlaTform) which acts like a DNA-powered computer chip. “These biological systems can heal if you design them correctly,” adds Adamala. “So you can imagine a computer that will eventually heal itself.”
The basics of biocomputing
Biocomputing and regular computing have many similarities. Like regular computing, biocomputing works by running information through a series of gates, usually logic gates. A logic gate works as a fork in the road for an electronic circuit. The input will travel one way or another, giving two different outputs. An example logic gate is the AND gate, which has two inputs (A and B) and two different results. If both A and B are 1, the AND gate output will be 1. If only A is 1 and B is 0, the output will be 0 and vice versa. If both A and B are 0, the result will be 0. While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output. In this case, the DNA enters the logic gate as a single or double strand.
If the DNA is double-stranded, the system “digests” the DNA or destroys it, which results in non-fluorescence or “0” output. Conversely, if the DNA is single-stranded, it won’t be digested and instead will be copied by several enzymes in the biocomputing system, resulting in fluorescent RNA or a “1” output. And the output for this type of binary system can be expanded beyond fluorescence or not. For example, a “1” output might be the production of the enzyme insulin, while a “0” may be that no insulin is produced. “This kind of synergy between biology and computation is the essence of biocomputing,” says Stephanie Forrest, a professor and the director of the Biodesign Center for Biocomputing, Security and Society at Arizona State University.
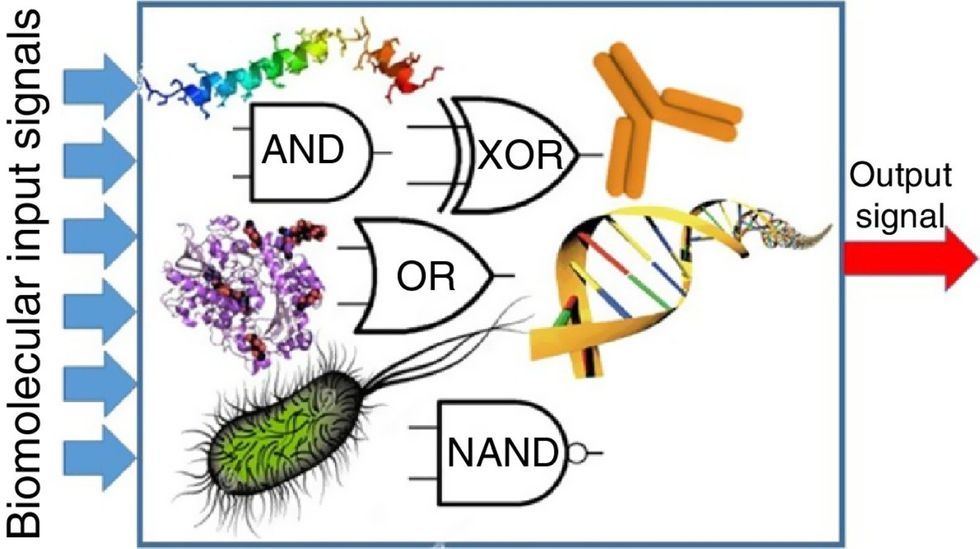
Biocomputing circles are made of DNA, RNA, proteins and even bacteria.
Evgeny Katz
The TRUMPET’s promise
Depending on whether the biocomputing system is placed directly inside a cell within the human body, or run in a test-tube, different environmental factors play a role. When an output is produced inside a cell, the cell's natural processes can amplify this output (for example, a specific protein or DNA strand), creating a solid signal. However, these cells can also be very leaky. “You want the cells to do the thing you ask them to do before they finish whatever their businesses, which is to grow, replicate, metabolize,” Adamala explains. “However, often the gate may be triggered without the right inputs, creating a false positive signal. So that's why natural logic gates are often leaky." While biocomputing outside a cell in a test tube can allow for tighter control over the logic gates, the outputs or signals cannot be amplified by a cell and are less potent.
TRUMPET, which is smaller than a cell, taps into both cellular and non-cellular biocomputing benefits. “At its core, it is a nonliving logic gate system,” Adamala states, “It's a DNA-based logic gate system. But because we use enzymes, and the readout is enzymatic [where an enzyme replicates the fluorescent RNA], we end up with signal amplification." This readout means that the output from the TRUMPET system, a fluorescent RNA strand, can be replicated by nearby enzymes in the platform, making the light signal stronger. "So it combines the best of both worlds,” Adamala adds.
These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body.
The TRUMPET biocomputing process is relatively straightforward. “If the DNA [input] shows up as single-stranded, it will not be digested [by the logic gate], and you get this nice fluorescent output as the RNA is made from the single-stranded DNA, and that's a 1,” Adamala explains. "And if the DNA input is double-stranded, it gets digested by the enzymes in the logic gate, and there is no RNA created from the DNA, so there is no fluorescence, and the output is 0." On the story's leading image above, if the tube is "lit" with a purple color, that is a binary 1 signal for computing. If it's "off" it is a 0.
While still in research, TRUMPET and other biocomputing systems promise significant benefits to personalized healthcare and medicine. These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body. The study’s lead author and graduate student Judee Sharon is already beginning to research TRUMPET's ability for earlier cancer diagnoses. Because the inputs for TRUMPET are single or double-stranded DNA, any mutated or cancerous DNA could theoretically be detected from the platform through the biocomputing process. Theoretically, devices like TRUMPET could be used to detect cancer and other diseases earlier.
Adamala sees TRUMPET not only as a detection system but also as a potential cancer drug delivery system. “Ideally, you would like the drug only to turn on when it senses the presence of a cancer cell. And that's how we use the logic gates, which work in response to inputs like cancerous DNA. Then the output can be the production of a small molecule or the release of a small molecule that can then go and kill what needs killing, in this case, a cancer cell. So we would like to develop applications that use this technology to control the logic gate response of a drug’s delivery to a cell.”
Although platforms like TRUMPET are making progress, a lot more work must be done before they can be used commercially. “The process of translating mechanisms and architecture from biology to computing and vice versa is still an art rather than a science,” says Forrest. “It requires deep computer science and biology knowledge,” she adds. “Some people have compared interdisciplinary science to fusion restaurants—not all combinations are successful, but when they are, the results are remarkable.”
Crickets are low on fat, high on protein, and can be farmed sustainably. They are also crunchy.
In today’s podcast episode, Leaps.org Deputy Editor Lina Zeldovich speaks about the health and ecological benefits of farming crickets for human consumption with Bicky Nguyen, who joins Lina from Vietnam. Bicky and her business partner Nam Dang operate an insect farm named CricketOne. Motivated by the idea of sustainable and healthy protein production, they started their unconventional endeavor a few years ago, despite numerous naysayers who didn’t believe that humans would ever consider munching on bugs.
Yet, making creepy crawlers part of our diet offers many health and planetary advantages. Food production needs to match the rise in global population, estimated to reach 10 billion by 2050. One challenge is that some of our current practices are inefficient, polluting and wasteful. According to nonprofit EarthSave.org, it takes 2,500 gallons of water, 12 pounds of grain, 35 pounds of topsoil and the energy equivalent of one gallon of gasoline to produce one pound of feedlot beef, although exact statistics vary between sources.
Meanwhile, insects are easy to grow, high on protein and low on fat. When roasted with salt, they make crunchy snacks. When chopped up, they transform into delicious pâtes, says Bicky, who invents her own cricket recipes and serves them at industry and public events. Maybe that’s why some research predicts that edible insects market may grow to almost $10 billion by 2030. Tune in for a delectable chat on this alternative and sustainable protein.
Listen on Apple | Listen on Spotify | Listen on Stitcher | Listen on Amazon | Listen on Google
Further reading:
More info on Bicky Nguyen
https://yseali.fulbright.edu.vn/en/faculty/bicky-n...
The environmental footprint of beef production
https://www.earthsave.org/environment.htm
https://www.watercalculator.org/news/articles/beef-king-big-water-footprints/
https://www.frontiersin.org/articles/10.3389/fsufs.2019.00005/full
https://ourworldindata.org/carbon-footprint-food-methane
Insect farming as a source of sustainable protein
https://www.insectgourmet.com/insect-farming-growing-bugs-for-protein/
https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/insect-farming
Cricket flour is taking the world by storm
https://www.cricketflours.com/
https://talk-commerce.com/blog/what-brands-use-cricket-flour-and-why/

Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.