DNA- and RNA-based electronic implants may revolutionize healthcare
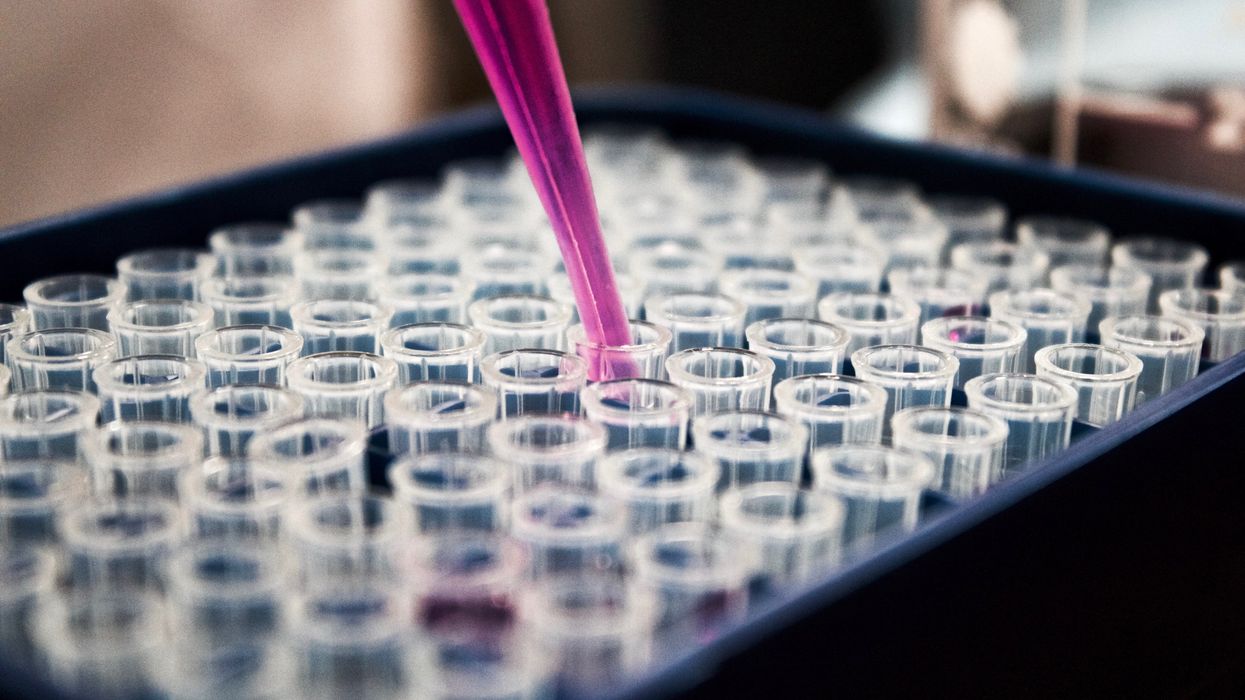
The test tubes contain tiny DNA/enzyme-based circuits, which comprise TRUMPET, a new type of electronic device, smaller than a cell.
Implantable electronic devices can significantly improve patients’ quality of life. A pacemaker can encourage the heart to beat more regularly. A neural implant, usually placed at the back of the skull, can help brain function and encourage higher neural activity. Current research on neural implants finds them helpful to patients with Parkinson’s disease, vision loss, hearing loss, and other nerve damage problems. Several of these implants, such as Elon Musk’s Neuralink, have already been approved by the FDA for human use.
Yet, pacemakers, neural implants, and other such electronic devices are not without problems. They require constant electricity, limited through batteries that need replacements. They also cause scarring. “The problem with doing this with electronics is that scar tissue forms,” explains Kate Adamala, an assistant professor of cell biology at the University of Minnesota Twin Cities. “Anytime you have something hard interacting with something soft [like muscle, skin, or tissue], the soft thing will scar. That's why there are no long-term neural implants right now.” To overcome these challenges, scientists are turning to biocomputing processes that use organic materials like DNA and RNA. Other promised benefits include “diagnostics and possibly therapeutic action, operating as nanorobots in living organisms,” writes Evgeny Katz, a professor of bioelectronics at Clarkson University, in his bookDNA- And RNA-Based Computing Systems.
While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output.
Adamala’s research focuses on developing such biocomputing systems using DNA, RNA, proteins, and lipids. Using these molecules in the biocomputing systems allows the latter to be biocompatible with the human body, resulting in a natural healing process. In a recent Nature Communicationsstudy, Adamala and her team created a new biocomputing platform called TRUMPET (Transcriptional RNA Universal Multi-Purpose GatE PlaTform) which acts like a DNA-powered computer chip. “These biological systems can heal if you design them correctly,” adds Adamala. “So you can imagine a computer that will eventually heal itself.”
The basics of biocomputing
Biocomputing and regular computing have many similarities. Like regular computing, biocomputing works by running information through a series of gates, usually logic gates. A logic gate works as a fork in the road for an electronic circuit. The input will travel one way or another, giving two different outputs. An example logic gate is the AND gate, which has two inputs (A and B) and two different results. If both A and B are 1, the AND gate output will be 1. If only A is 1 and B is 0, the output will be 0 and vice versa. If both A and B are 0, the result will be 0. While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output. In this case, the DNA enters the logic gate as a single or double strand.
If the DNA is double-stranded, the system “digests” the DNA or destroys it, which results in non-fluorescence or “0” output. Conversely, if the DNA is single-stranded, it won’t be digested and instead will be copied by several enzymes in the biocomputing system, resulting in fluorescent RNA or a “1” output. And the output for this type of binary system can be expanded beyond fluorescence or not. For example, a “1” output might be the production of the enzyme insulin, while a “0” may be that no insulin is produced. “This kind of synergy between biology and computation is the essence of biocomputing,” says Stephanie Forrest, a professor and the director of the Biodesign Center for Biocomputing, Security and Society at Arizona State University.
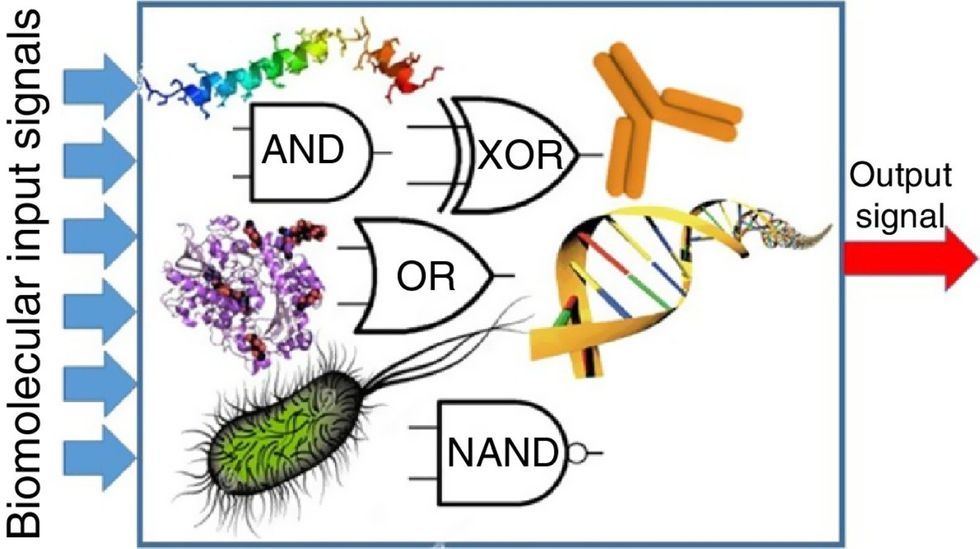
Biocomputing circles are made of DNA, RNA, proteins and even bacteria.
Evgeny Katz
The TRUMPET’s promise
Depending on whether the biocomputing system is placed directly inside a cell within the human body, or run in a test-tube, different environmental factors play a role. When an output is produced inside a cell, the cell's natural processes can amplify this output (for example, a specific protein or DNA strand), creating a solid signal. However, these cells can also be very leaky. “You want the cells to do the thing you ask them to do before they finish whatever their businesses, which is to grow, replicate, metabolize,” Adamala explains. “However, often the gate may be triggered without the right inputs, creating a false positive signal. So that's why natural logic gates are often leaky." While biocomputing outside a cell in a test tube can allow for tighter control over the logic gates, the outputs or signals cannot be amplified by a cell and are less potent.
TRUMPET, which is smaller than a cell, taps into both cellular and non-cellular biocomputing benefits. “At its core, it is a nonliving logic gate system,” Adamala states, “It's a DNA-based logic gate system. But because we use enzymes, and the readout is enzymatic [where an enzyme replicates the fluorescent RNA], we end up with signal amplification." This readout means that the output from the TRUMPET system, a fluorescent RNA strand, can be replicated by nearby enzymes in the platform, making the light signal stronger. "So it combines the best of both worlds,” Adamala adds.
These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body.
The TRUMPET biocomputing process is relatively straightforward. “If the DNA [input] shows up as single-stranded, it will not be digested [by the logic gate], and you get this nice fluorescent output as the RNA is made from the single-stranded DNA, and that's a 1,” Adamala explains. "And if the DNA input is double-stranded, it gets digested by the enzymes in the logic gate, and there is no RNA created from the DNA, so there is no fluorescence, and the output is 0." On the story's leading image above, if the tube is "lit" with a purple color, that is a binary 1 signal for computing. If it's "off" it is a 0.
While still in research, TRUMPET and other biocomputing systems promise significant benefits to personalized healthcare and medicine. These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body. The study’s lead author and graduate student Judee Sharon is already beginning to research TRUMPET's ability for earlier cancer diagnoses. Because the inputs for TRUMPET are single or double-stranded DNA, any mutated or cancerous DNA could theoretically be detected from the platform through the biocomputing process. Theoretically, devices like TRUMPET could be used to detect cancer and other diseases earlier.
Adamala sees TRUMPET not only as a detection system but also as a potential cancer drug delivery system. “Ideally, you would like the drug only to turn on when it senses the presence of a cancer cell. And that's how we use the logic gates, which work in response to inputs like cancerous DNA. Then the output can be the production of a small molecule or the release of a small molecule that can then go and kill what needs killing, in this case, a cancer cell. So we would like to develop applications that use this technology to control the logic gate response of a drug’s delivery to a cell.”
Although platforms like TRUMPET are making progress, a lot more work must be done before they can be used commercially. “The process of translating mechanisms and architecture from biology to computing and vice versa is still an art rather than a science,” says Forrest. “It requires deep computer science and biology knowledge,” she adds. “Some people have compared interdisciplinary science to fusion restaurants—not all combinations are successful, but when they are, the results are remarkable.”
How the Human Brain Project Built a Mind of its Own
In 2013, the Human Brain Project set out to build a realistic computer model of the brain over ten years. Now, experts are reflecting on HBP's achievements with an eye toward the future.
In 2009, neuroscientist Henry Markram gave an ambitious TED talk. “Our mission is to build a detailed, realistic computer model of the human brain,” he said, naming three reasons for this unmatched feat of engineering. One was because understanding the human brain was essential to get along in society. Another was because experimenting on animal brains could only get scientists so far in understanding the human ones. Third, medicines for mental disorders weren’t good enough. “There are two billion people on the planet that are affected by mental disorders, and the drugs that are used today are largely empirical,” Markram said. “I think that we can come up with very concrete solutions on how to treat disorders.”
Markram's arguments were very persuasive. In 2013, the European Commission launched the Human Brain Project, or HBP, as part of its Future and Emerging Technologies program. Viewed as Europe’s chance to try to win the “brain race” between the U.S., China, Japan, and other countries, the project received about a billion euros in funding with the goal to simulate the entire human brain on a supercomputer, or in silico, by 2023.
Now, after 10 years of dedicated neuroscience research, the HBP is coming to an end. As its many critics warned, it did not manage to build an entire human brain in silico. Instead, it achieved a multifaceted array of different goals, some of them unexpected.
Scholars have found that the project did help advance neuroscience more than some detractors initially expected, specifically in the area of brain simulations and virtual models. Using an interdisciplinary approach of combining technology, such as AI and digital simulations, with neuroscience, the HBP worked to gain a deeper understanding of the human brain’s complicated structure and functions, which in some cases led to novel treatments for brain disorders. Lastly, through online platforms, the HBP spearheaded a previously unmatched level of global neuroscience collaborations.
Simulating a human brain stirs up controversy
Right from the start, the project was plagued with controversy and condemnation. One of its prominent critics was Yves Fregnac, a professor in cognitive science at the Polytechnic Institute of Paris and research director at the French National Centre for Scientific Research. Fregnac argued in numerous articles that the HBP was overfunded based on proposals with unrealistic goals. “This new way of over-selling scientific targets, deeply aligned with what modern society expects from mega-sciences in the broad sense (big investment, big return), has been observed on several occasions in different scientific sub-fields,” he wrote in one of his articles, “before invading the field of brain sciences and neuromarketing.”
"A human brain model can simulate an experiment a million times for many different conditions, but the actual human experiment can be performed only once or a few times," said Viktor Jirsa, a professor at Aix-Marseille University.
Responding to such critiques, the HBP worked to restructure the effort in its early days with new leadership, organization, and goals that were more flexible and attainable. “The HBP got a more versatile, pluralistic approach,” said Viktor Jirsa, a professor at Aix-Marseille University and one of the HBP lead scientists. He believes that these changes fixed at least some of HBP’s issues. “The project has been on a very productive and scientifically fruitful course since then.”
After restructuring, the HBP became a European hub on brain research, with hundreds of scientists joining its growing network. The HBP created projects focused on various brain topics, from consciousness to neurodegenerative diseases. HBP scientists worked on complex subjects, such as mapping out the brain, combining neuroscience and robotics, and experimenting with neuromorphic computing, a computational technique inspired by the human brain structure and function—to name just a few.
Simulations advance knowledge and treatment options
In 2013, it seemed that bringing neuroscience into a digital age would be farfetched, but research within the HBP has made this achievable. The virtual maps and simulations various HBP teams create through brain imaging data make it easier for neuroscientists to understand brain developments and functions. The teams publish these models on the HBP’s EBRAINS online platform—one of the first to offer access to such data to neuroscientists worldwide via an open-source online site. “This digital infrastructure is backed by high-performance computers, with large datasets and various computational tools,” said Lucy Xiaolu Wang, an assistant professor in the Resource Economics Department at the University of Massachusetts Amherst, who studies the economics of the HBP. That means it can be used in place of many different types of human experimentation.
Jirsa’s team is one of many within the project that works on virtual brain models and brain simulations. Compiling patient data, Jirsa and his team can create digital simulations of different brain activities—and repeat these experiments many times, which isn’t often possible in surgeries on real brains. “A human brain model can simulate an experiment a million times for many different conditions,” Jirsa explained, “but the actual human experiment can be performed only once or a few times.” Using simulations also saves scientists and doctors time and money when looking at ways to diagnose and treat patients with brain disorders.

Compiling patient data, scientists can create digital simulations of different brain activities—and repeat these experiments many times.
The Human Brain Project
Simulations can help scientists get a full picture that otherwise is unattainable. “Another benefit is data completion,” added Jirsa, “in which incomplete data can be complemented by the model. In clinical settings, we can often measure only certain brain areas, but when linked to the brain model, we can enlarge the range of accessible brain regions and make better diagnostic predictions.”
With time, Jirsa’s team was able to move into patient-specific simulations. “We advanced from generic brain models to the ability to use a specific patient’s brain data, from measurements like MRI and others, to create individualized predictive models and simulations,” Jirsa explained. He and his team are working on this personalization technique to treat patients with epilepsy. According to the World Health Organization, about 50 million people worldwide suffer from epilepsy, a disorder that causes recurring seizures. While some epilepsy causes are known others remain an enigma, and many are hard to treat. For some patients whose epilepsy doesn’t respond to medications, removing part of the brain where seizures occur may be the only option. Understanding where in the patients’ brains seizures arise can give scientists a better idea of how to treat them and whether to use surgery versus medications.
“We apply such personalized models…to precisely identify where in a patient’s brain seizures emerge,” Jirsa explained. “This guides individual surgery decisions for patients for which surgery is the only treatment option.” He credits the HBP for the opportunity to develop this novel approach. “The personalization of our epilepsy models was only made possible by the Human Brain Project, in which all the necessary tools have been developed. Without the HBP, the technology would not be in clinical trials today.”
Personalized simulations can significantly advance treatments, predict the outcome of specific medical procedures and optimize them before actually treating patients. Jirsa is watching this happen firsthand in his ongoing research. “Our technology for creating personalized brain models is now used in a large clinical trial for epilepsy, funded by the French state, where we collaborate with clinicians in hospitals,” he explained. “We have also founded a spinoff company called VB Tech (Virtual Brain Technologies) to commercialize our personalized brain model technology and make it available to all patients.”
The Human Brain Project created a level of interconnectedness within the neuroscience research community that never existed before—a network not unlike the brain’s own.
Other experts believe it’s too soon to tell whether brain simulations could change epilepsy treatments. “The life cycle of developing treatments applicable to patients often runs over a decade,” Wang stated. “It is still too early to draw a clear link between HBP’s various project areas with patient care.” However, she admits that some studies built on the HBP-collected knowledge are already showing promise. “Researchers have used neuroscientific atlases and computational tools to develop activity-specific stimulation programs that enabled paraplegic patients to move again in a small-size clinical trial,” Wang said. Another intriguing study looked at simulations of Alzheimer’s in the brain to understand how it evolves over time.
Some challenges remain hard to overcome even with computer simulations. “The major challenge has always been the parameter explosion, which means that many different model parameters can lead to the same result,” Jirsa explained. An example of this parameter explosion could be two different types of neurodegenerative conditions, such as Parkinson’s and Huntington’s diseases. Both afflict the same area of the brain, the basal ganglia, which can affect movement, but are caused by two different underlying mechanisms. “We face the same situation in the living brain, in which a large range of diverse mechanisms can produce the same behavior,” Jirsa said. The simulations still have to overcome the same challenge.
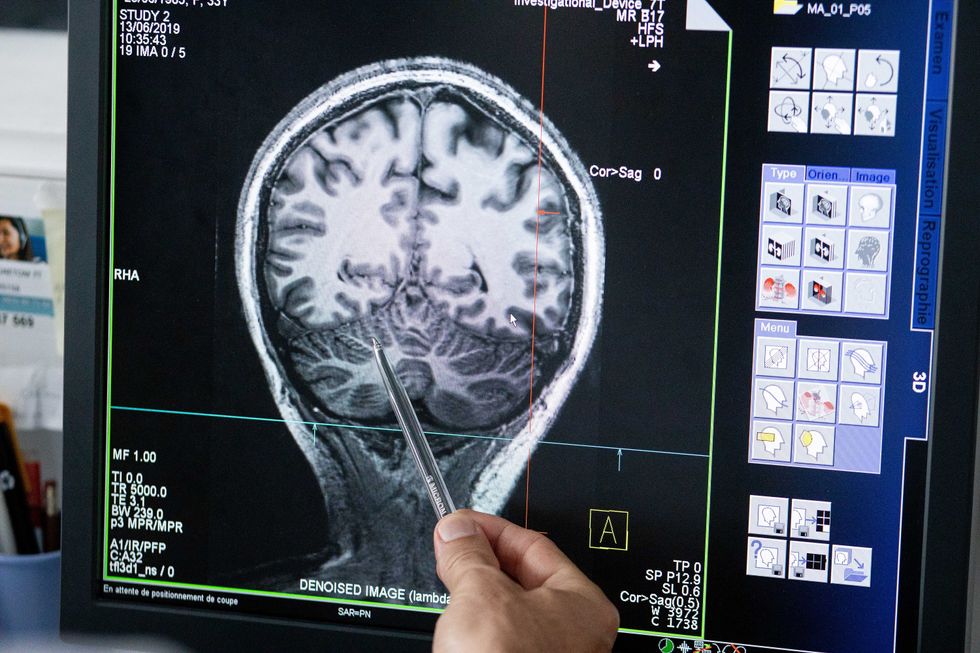
Understanding where in the patients’ brains seizures arise can give scientists a better idea of how to treat them and whether to use surgery versus medications.
The Human Brain Project
A network not unlike the brain’s own
Though the HBP will be closing this year, its legacy continues in various studies, spin-off companies, and its online platform, EBRAINS. “The HBP is one of the earliest brain initiatives in the world, and the 10-year long-term goal has united many researchers to collaborate on brain sciences with advanced computational tools,” Wang said. “Beyond the many research articles and projects collaborated on during the HBP, the online neuroscience research infrastructure EBRAINS will be left as a legacy even after the project ends.”
Those who worked within the HBP see the end of this project as the next step in neuroscience research. “Neuroscience has come closer to very meaningful applications through the systematic link with new digital technologies and collaborative work,” Jirsa stated. “In that way, the project really had a pioneering role.” It also created a level of interconnectedness within the neuroscience research community that never existed before—a network not unlike the brain’s own. “Interconnectedness is an important advance and prerequisite for progress,” Jirsa said. “The neuroscience community has in the past been rather fragmented and this has dramatically changed in recent years thanks to the Human Brain Project.”
According to its website, by 2023 HBP’s network counted over 500 scientists from over 123 institutions and 16 different countries, creating one of the largest multi-national research groups in the world. Even though the project hasn’t produced the in-silico brain as Markram envisioned it, the HBP created a communal mind with immense potential. “It has challenged us to think beyond the boundaries of our own laboratories,” Jirsa said, “and enabled us to go much further together than we could have ever conceived going by ourselves.”