Scientists redesign bacteria to tackle the antibiotic resistance crisis

Probiotic bacteria can be engineered to fight antibiotic-resistant superbugs by releasing chemicals that kill them.
In 1945, almost two decades after Alexander Fleming discovered penicillin, he warned that as antibiotics use grows, they may lose their efficiency. He was prescient—the first case of penicillin resistance was reported two years later. Back then, not many people paid attention to Fleming’s warning. After all, the “golden era” of the antibiotics age had just began. By the 1950s, three new antibiotics derived from soil bacteria — streptomycin, chloramphenicol, and tetracycline — could cure infectious diseases like tuberculosis, cholera, meningitis and typhoid fever, among others.
Today, these antibiotics and many of their successors developed through the 1980s are gradually losing their effectiveness. The extensive overuse and misuse of antibiotics led to the rise of drug resistance. The livestock sector buys around 80 percent of all antibiotics sold in the U.S. every year. Farmers feed cows and chickens low doses of antibiotics to prevent infections and fatten up the animals, which eventually causes resistant bacterial strains to evolve. If manure from cattle is used on fields, the soil and vegetables can get contaminated with antibiotic-resistant bacteria. Another major factor is doctors overprescribing antibiotics to humans, particularly in low-income countries. Between 2000 to 2018, the global rates of human antibiotic consumption shot up by 46 percent.
In recent years, researchers have been exploring a promising avenue: the use of synthetic biology to engineer new bacteria that may work better than antibiotics. The need continues to grow, as a Lancet study linked antibiotic resistance to over 1.27 million deaths worldwide in 2019, surpassing HIV/AIDS and malaria. The western sub-Saharan Africa region had the highest death rate (27.3 people per 100,000).
Researchers warn that if nothing changes, by 2050, antibiotic resistance could kill 10 million people annually.
To make it worse, our remedy pipelines are drying up. Out of the 18 biggest pharmaceutical companies, 15 abandoned antibiotic development by 2013. According to the AMR Action Fund, venture capital has remained indifferent towards biotech start-ups developing new antibiotics. In 2019, at least two antibiotic start-ups filed for bankruptcy. As of December 2020, there were 43 new antibiotics in clinical development. But because they are based on previously known molecules, scientists say they are inadequate for treating multidrug-resistant bacteria. Researchers warn that if nothing changes, by 2050, antibiotic resistance could kill 10 million people annually.
The rise of synthetic biology
To circumvent this dire future, scientists have been working on alternative solutions using synthetic biology tools, meaning genetically modifying good bacteria to fight the bad ones.
From the time life evolved on earth around 3.8 billion years ago, bacteria have engaged in biological warfare. They constantly strategize new methods to combat each other by synthesizing toxic proteins that kill competition.
For example, Escherichia coli produces bacteriocins or toxins to kill other strains of E.coli that attempt to colonize the same habitat. Microbes like E.coli (which are not all pathogenic) are also naturally present in the human microbiome. The human microbiome harbors up to 100 trillion symbiotic microbial cells. The majority of them are beneficial organisms residing in the gut at different compositions.
The chemicals that these “good bacteria” produce do not pose any health risks to us, but can be toxic to other bacteria, particularly to human pathogens. For the last three decades, scientists have been manipulating bacteria’s biological warfare tactics to our collective advantage.
In the late 1990s, researchers drew inspiration from electrical and computing engineering principles that involve constructing digital circuits to control devices. In certain ways, every cell in living organisms works like a tiny computer. The cell receives messages in the form of biochemical molecules that cling on to its surface. Those messages get processed within the cells through a series of complex molecular interactions.
Synthetic biologists can harness these living cells’ information processing skills and use them to construct genetic circuits that perform specific instructions—for example, secrete a toxin that kills pathogenic bacteria. “Any synthetic genetic circuit is merely a piece of information that hangs around in the bacteria’s cytoplasm,” explains José Rubén Morones-Ramírez, a professor at the Autonomous University of Nuevo León, Mexico. Then the ribosome, which synthesizes proteins in the cell, processes that new information, making the compounds scientists want bacteria to make. “The genetic circuit remains separated from the living cell’s DNA,” Morones-Ramírez explains. When the engineered bacteria replicates, the genetic circuit doesn’t become part of its genome.
Highly intelligent by bacterial standards, some multidrug resistant V. cholerae strains can also “collaborate” with other intestinal bacterial species to gain advantage and take hold of the gut.
In 2000, Boston-based researchers constructed an E.coli with a genetic switch that toggled between turning genes on and off two. Later, they built some safety checks into their bacteria. “To prevent unintentional or deleterious consequences, in 2009, we built a safety switch in the engineered bacteria’s genetic circuit that gets triggered after it gets exposed to a pathogen," says James Collins, a professor of biological engineering at MIT and faculty member at Harvard University’s Wyss Institute. “After getting rid of the pathogen, the engineered bacteria is designed to switch off and leave the patient's body.”

Overuse and misuse of antibiotics causes resistant strains to evolve
Adobe Stock
Seek and destroy
As the field of synthetic biology developed, scientists began using engineered bacteria to tackle superbugs. They first focused on Vibrio cholerae, which in the 19th and 20th century caused cholera pandemics in India, China, the Middle East, Europe, and Americas. Like many other bacteria, V. cholerae communicate with each other via quorum sensing, a process in which the microorganisms release different signaling molecules, to convey messages to its brethren. Highly intelligent by bacterial standards, some multidrug resistant V. cholerae strains can also “collaborate” with other intestinal bacterial species to gain advantage and take hold of the gut. When untreated, cholera has a mortality rate of 25 to 50 percent and outbreaks frequently occur in developing countries, especially during floods and droughts.
Sometimes, however, V. cholerae makes mistakes. In 2008, researchers at Cornell University observed that when quorum sensing V. cholerae accidentally released high concentrations of a signaling molecule called CAI-1, it had a counterproductive effect—the pathogen couldn’t colonize the gut.
So the group, led by John March, professor of biological and environmental engineering, developed a novel strategy to combat V. cholerae. They genetically engineered E.coli to eavesdrop on V. cholerae communication networks and equipped it with the ability to release the CAI-1 molecules. That interfered with V. cholerae progress. Two years later, the Cornell team showed that V. cholerae-infected mice treated with engineered E.coli had a 92 percent survival rate.
These findings inspired researchers to sic the good bacteria present in foods like yogurt and kimchi onto the drug-resistant ones.
Three years later in 2011, Singapore-based scientists engineered E.coli to detect and destroy Pseudomonas aeruginosa, an often drug-resistant pathogen that causes pneumonia, urinary tract infections, and sepsis. Once the genetically engineered E.coli found its target through its quorum sensing molecules, it then released a peptide, that could eradicate 99 percent of P. aeruginosa cells in a test-tube experiment. The team outlined their work in a Molecular Systems Biology study.
“At the time, we knew that we were entering new, uncharted territory,” says lead author Matthew Chang, an associate professor and synthetic biologist at the National University of Singapore and lead author of the study. “To date, we are still in the process of trying to understand how long these microbes stay in our bodies and how they might continue to evolve.”
More teams followed the same path. In a 2013 study, MIT researchers also genetically engineered E.coli to detect P. aeruginosa via the pathogen’s quorum-sensing molecules. It then destroyed the pathogen by secreting a lab-made toxin.
Probiotics that fight
A year later in 2014, a Nature study found that the abundance of Ruminococcus obeum, a probiotic bacteria naturally occurring in the human microbiome, interrupts and reduces V.cholerae’s colonization— by detecting the pathogen’s quorum sensing molecules. The natural accumulation of R. obeum in Bangladeshi adults helped them recover from cholera despite living in an area with frequent outbreaks.
The findings from 2008 to 2014 inspired Collins and his team to delve into how good bacteria present in foods like yogurt and kimchi can attack drug-resistant bacteria. In 2018, Collins and his team developed the engineered probiotic strategy. They tweaked a bacteria commonly found in yogurt called Lactococcus lactis to treat cholera.
Engineered bacteria can be trained to target pathogens when they are at their most vulnerable metabolic stage in the human gut. --José Rubén Morones-Ramírez.
More scientists followed with more experiments. So far, researchers have engineered various probiotic organisms to fight pathogenic bacteria like Staphylococcus aureus (leading cause of skin, tissue, bone, joint and blood infections) and Clostridium perfringens (which causes watery diarrhea) in test-tube and animal experiments. In 2020, Russian scientists engineered a probiotic called Pichia pastoris to produce an enzyme called lysostaphin that eradicated S. aureus in vitro. Another 2020 study from China used an engineered probiotic bacteria Lactobacilli casei as a vaccine to prevent C. perfringens infection in rabbits.
In a study last year, Ramírez’s group at the Autonomous University of Nuevo León, engineered E. coli to detect quorum-sensing molecules from Methicillin-resistant Staphylococcus aureus or MRSA, a notorious superbug. The E. coli then releases a bacteriocin that kills MRSA. “An antibiotic is just a molecule that is not intelligent,” says Ramírez. “On the other hand, engineered bacteria can be trained to target pathogens when they are at their most vulnerable metabolic stage in the human gut.”
Collins and Timothy Lu, an associate professor of biological engineering at MIT, found that engineered E. coli can help treat other conditions—such as phenylketonuria, a rare metabolic disorder, that causes the build-up of an amino acid phenylalanine. Their start-up Synlogic aims to commercialize the technology, and has completed a phase 2 clinical trial.
Circumventing the challenges
The bacteria-engineering technique is not without pitfalls. One major challenge is that beneficial gut bacteria produce their own quorum-sensing molecules that can be similar to those that pathogens secrete. If an engineered bacteria’s biosensor is not specific enough, it will be ineffective.
Another concern is whether engineered bacteria might mutate after entering the gut. “As with any technology, there are risks where bad actors could have the capability to engineer a microbe to act quite nastily,” says Collins of MIT. But Collins and Ramírez both insist that the chances of the engineered bacteria mutating on its own are virtually non-existent. “It is extremely unlikely for the engineered bacteria to mutate,” Ramírez says. “Coaxing a living cell to do anything on command is immensely challenging. Usually, the greater risk is that the engineered bacteria entirely lose its functionality.”
However, the biggest challenge is bringing the curative bacteria to consumers. Pharmaceutical companies aren’t interested in antibiotics or their alternatives because it’s less profitable than developing new medicines for non-infectious diseases. Unlike the more chronic conditions like diabetes or cancer that require long-term medications, infectious diseases are usually treated much quicker. Running clinical trials are expensive and antibiotic-alternatives aren’t lucrative enough.
“Unfortunately, new medications for antibiotic resistant infections have been pushed to the bottom of the field,” says Lu of MIT. “It's not because the technology does not work. This is more of a market issue. Because clinical trials cost hundreds of millions of dollars, the only solution is that governments will need to fund them.” Lu stresses that societies must lobby to change how the modern healthcare industry works. “The whole world needs better treatments for antibiotic resistance.”
7 Reasons Why We Should Not Need Boosters for COVID-19
A top infectious disease physician explains why immunity derived from natural infection and the vaccines should be long-lasting.
There are at least 7 reasons why immunity after vaccination or infection with COVID-19 should likely be long-lived. If durable, I do not think boosters will be necessary in the future, despite CEOs of pharmaceutical companies (who stand to profit from boosters) messaging that they may and readying such boosters. To explain these reasons, let's orient ourselves to the main components of the immune system.
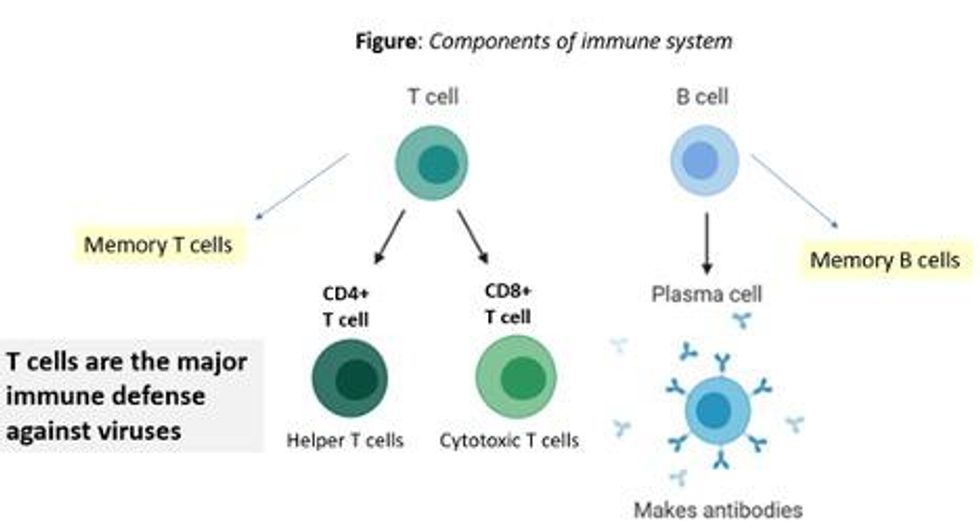
There are two major arms of the immune system: B cells (which produce antibodies) and T cells (which are formed specifically to attack and kill pathogens). T cells are divided into two types, CD4 cells ("helper" T cells) and CD8 cells ("cytotoxic" T cells).
Each arm, once stimulated by infection or vaccine, should hopefully make "memory" banks. So if the body sees the pathogen in the future, these defenses should come roaring back to attack the virus and protect you from getting sick. Plenty of research in COVID-19 indicates a likely long-lasting response to the vaccine or infection. Here are seven of the most compelling reasons:
REASON 1: Memory B Cells Are Produced By Vaccines and Natural Infection
In one study, 12 volunteers who had never had Covid-19--and were fully vaccinated with two Pfizer/BioNTech shots-- underwent biopsies of their lymph nodes. This is where memory B cells are stored in places called "germinal centers". The biopsies were performed three, four, six, and seven weeks after the first mRNA vaccine shot, and were stained to reveal that germinal center memory B cells in the lymph nodes increased in concentration over time.
Natural infection also generates memory B cells. Even after antibody levels wane over time, strong memory B cells were detected in the blood of individuals six and eight months after infection in different studies. Indeed, the half-lives of the memory B cells seen in the study examining patients 8 months after COVID-19 led the authors to conclude that "B cell memory to SARS-CoV-2 was robust and is likely long-lasting." Reason #2 tells us that memory B cells can be active for a very long time indeed.
REASON #2: Memory B Cells Can Produce Neutralizing Antibodies If They See Infection Again Decades Later
Demonstrated production of memory B cells after vaccination or natural infection with COVID-19 is so important because memory B cells, once generated, can be activated to produce high levels of neutralizing antibodies against the pathogen even if encountered many years after the initial exposure. In one amazing study (published in 2008), researchers isolated memory B cells against the 1918 flu strain from the blood of 32 individuals aged 91-101 years. These people had been born on or before 1915 and had survived that pandemic.
Their memory B cells, when exposed to the 1918 flu strain in a test tube, generated high levels of neutralizing antibodies against the virus -- antibodies that then protected mice from lethal infection with this deadly strain. The ability of memory B cells to produce complex antibody responses against an infection nine decades after exposure speaks to their durability.
REASON #3: Vaccines or Natural Infection Trigger Strong Memory T Cell Immunity
All of the trials of the major COVID-19 vaccine candidates measured strong T cell immunity following vaccination, most often assessed by measuring SARS-CoV-2 specific T cells in the phase I/II safety and immunogenicity studies. There are a number of studies that demonstrate the production of strong T cell immunity to COVID-19 after natural infection as well, even when the infection was mild or asymptomatic.
The same study that showed us robust memory B cell production 8 months after natural infection also demonstrated strong and sustained memory T cell production. In fact, the half-lives of the memory T cells in this cohort were long (~125-225 days for CD8+ and ~94-153 days for CD4+ T cells), comparable to the 123-day half-life observed for memory CD8+ T cells after yellow fever immunization (a vaccine usually given once over a lifetime).
A recent study of individuals recovered from COVID-19 show that the initial T cells generated by natural infection mature and differentiate over time into memory T cells that will be "put in the bank" for sustained periods.
REASON #4: T Cell Immunity Following Vaccinations for Other Infections Is Long-Lasting
Last year, we were fortunate to be able to measure how T cell immunity is generated by COVID-19 vaccines, which was not possible in earlier eras when vaccine trials were done for other infections (such as measles, mumps, rubella, pertussis, diphtheria). Antibodies are just the "tip of the iceberg" when assessing the response to vaccination, but were the only arm of the immune response that could be measured following vaccination in the past.
Measuring pathogen-specific T cell responses takes sophisticated technology. However, T cell responses, when assessed years after vaccination for other pathogens, has been shown to be long-lasting. For example, in one study of 56 volunteers who had undergone measles vaccination when they were much younger, strong CD8 and CD4 cell responses to vaccination could be detected up to 34 years later.
REASON #5: T Cell Immunity to Related Coronaviruses That Caused Severe Disease is Long-Lasting
SARS-CoV-2 is a coronavirus that causes severe disease, unlike coronaviruses that cause the common cold. Two other coronaviruses in the recent past caused severe disease, specifically Severely Acute Respiratory Distress Syndrome (SARS) in late 2002-2003 and Middle East Respiratory Syndrome (MERS) in 2011.
A study performed in 2020 demonstrated that the blood of 23 recovered SARS patients possess long-lasting memory T cells that were still reactive to SARS 17 years after the outbreak in 2003. Many scientists expect that T cell immunity to SARS-CoV-2 will be equally durable to that of its cousin.
REASON #6: T Cell Responses from Vaccination and Natural Infection With the Ancestral Strain of COVID-19 Are Robust Against Variants
Even though antibody responses from vaccination may be slightly lower against various COVID-19 variants of concern that have emerged in recent months, T cell immunity after vaccination has been shown to be unperturbed by mutations in the spike protein (in the variants). For instance, T cell responses after mRNA vaccines maintained strong activity against different variants (including P.1 Brazil variant, B.1.1.7 UK variant, B.1.351 South Africa variant and the CA.20.C California variant) in a recent study.
Another study showed that the vaccines generated robust T cell immunity that was unfazed by different variants, including B.1.351 and B.1.1.7. The CD4 and CD8 responses generated after natural infection are equally robust, showing activity against multiple "epitopes" (little segments) of the spike protein of the virus. For instance, CD8 cells responds to 52 epitopes and CD4 cells respond to 57 epitopes across the spike protein, so that a few mutations in the variants cannot knock out such a robust and in-breadth T cell response. Indeed, a recent paper showed that mRNA vaccines were 97.4 percent effective against severe COVID-19 disease in Qatar, even when the majority of circulating virus there was from variants of concern (B.1.351 and B.1.1.7).
REASON #7: Coronaviruses Don't Mutate Quickly Like Influenza, Which Requires Annual Booster Shots
Coronaviruses are RNA viruses, like influenza and HIV (which is actually a retrovirus), but do not mutate as quickly as either one. The reason that coronaviruses don't mutate very rapidly is that their replicating mechanism (polymerase) has a strong proofreading mechanism: If the virus mutates, it usually goes back and self-corrects. Mutations can arise with high rates of replication when transmission is very frequent -- as has been seen in recent months with the emergence of SARS-CoV-2 variants during surges. However, the COVID-19 virus will not be mutating like this when we tamp down transmission with mass vaccination.
In conclusion, I and many of my infectious disease colleagues expect the immunity from natural infection or vaccination to COVID-19 to be durable. Let's put discussion of boosters aside and work hard on global vaccine equity and distribution since the pandemic is not over until it is over for us all.
Professor Tim Caulfield stops by the "Making Sense of Science" podcast this month to discuss the infodemic around COVID-19 and how to deal with it.
The "Making Sense of Science" podcast features interviews with leading medical and scientific experts about the latest developments and the big ethical and societal questions they raise. This monthly podcast is hosted by journalist Kira Peikoff, founding editor of the award-winning science outlet Leaps.org.
Hear the 30-second trailer:
Listen to the whole episode:
.
Kira Peikoff was the editor-in-chief of Leaps.org from 2017 to 2021. As a journalist, her work has appeared in The New York Times, Newsweek, Nautilus, Popular Mechanics, The New York Academy of Sciences, and other outlets. She is also the author of four suspense novels that explore controversial issues arising from scientific innovation: Living Proof, No Time to Die, Die Again Tomorrow, and Mother Knows Best. Peikoff holds a B.A. in Journalism from New York University and an M.S. in Bioethics from Columbia University. She lives in New Jersey with her husband and two young sons. Follow her on Twitter @KiraPeikoff.

