Scientists redesign bacteria to tackle the antibiotic resistance crisis

Probiotic bacteria can be engineered to fight antibiotic-resistant superbugs by releasing chemicals that kill them.
In 1945, almost two decades after Alexander Fleming discovered penicillin, he warned that as antibiotics use grows, they may lose their efficiency. He was prescient—the first case of penicillin resistance was reported two years later. Back then, not many people paid attention to Fleming’s warning. After all, the “golden era” of the antibiotics age had just began. By the 1950s, three new antibiotics derived from soil bacteria — streptomycin, chloramphenicol, and tetracycline — could cure infectious diseases like tuberculosis, cholera, meningitis and typhoid fever, among others.
Today, these antibiotics and many of their successors developed through the 1980s are gradually losing their effectiveness. The extensive overuse and misuse of antibiotics led to the rise of drug resistance. The livestock sector buys around 80 percent of all antibiotics sold in the U.S. every year. Farmers feed cows and chickens low doses of antibiotics to prevent infections and fatten up the animals, which eventually causes resistant bacterial strains to evolve. If manure from cattle is used on fields, the soil and vegetables can get contaminated with antibiotic-resistant bacteria. Another major factor is doctors overprescribing antibiotics to humans, particularly in low-income countries. Between 2000 to 2018, the global rates of human antibiotic consumption shot up by 46 percent.
In recent years, researchers have been exploring a promising avenue: the use of synthetic biology to engineer new bacteria that may work better than antibiotics. The need continues to grow, as a Lancet study linked antibiotic resistance to over 1.27 million deaths worldwide in 2019, surpassing HIV/AIDS and malaria. The western sub-Saharan Africa region had the highest death rate (27.3 people per 100,000).
Researchers warn that if nothing changes, by 2050, antibiotic resistance could kill 10 million people annually.
To make it worse, our remedy pipelines are drying up. Out of the 18 biggest pharmaceutical companies, 15 abandoned antibiotic development by 2013. According to the AMR Action Fund, venture capital has remained indifferent towards biotech start-ups developing new antibiotics. In 2019, at least two antibiotic start-ups filed for bankruptcy. As of December 2020, there were 43 new antibiotics in clinical development. But because they are based on previously known molecules, scientists say they are inadequate for treating multidrug-resistant bacteria. Researchers warn that if nothing changes, by 2050, antibiotic resistance could kill 10 million people annually.
The rise of synthetic biology
To circumvent this dire future, scientists have been working on alternative solutions using synthetic biology tools, meaning genetically modifying good bacteria to fight the bad ones.
From the time life evolved on earth around 3.8 billion years ago, bacteria have engaged in biological warfare. They constantly strategize new methods to combat each other by synthesizing toxic proteins that kill competition.
For example, Escherichia coli produces bacteriocins or toxins to kill other strains of E.coli that attempt to colonize the same habitat. Microbes like E.coli (which are not all pathogenic) are also naturally present in the human microbiome. The human microbiome harbors up to 100 trillion symbiotic microbial cells. The majority of them are beneficial organisms residing in the gut at different compositions.
The chemicals that these “good bacteria” produce do not pose any health risks to us, but can be toxic to other bacteria, particularly to human pathogens. For the last three decades, scientists have been manipulating bacteria’s biological warfare tactics to our collective advantage.
In the late 1990s, researchers drew inspiration from electrical and computing engineering principles that involve constructing digital circuits to control devices. In certain ways, every cell in living organisms works like a tiny computer. The cell receives messages in the form of biochemical molecules that cling on to its surface. Those messages get processed within the cells through a series of complex molecular interactions.
Synthetic biologists can harness these living cells’ information processing skills and use them to construct genetic circuits that perform specific instructions—for example, secrete a toxin that kills pathogenic bacteria. “Any synthetic genetic circuit is merely a piece of information that hangs around in the bacteria’s cytoplasm,” explains José Rubén Morones-Ramírez, a professor at the Autonomous University of Nuevo León, Mexico. Then the ribosome, which synthesizes proteins in the cell, processes that new information, making the compounds scientists want bacteria to make. “The genetic circuit remains separated from the living cell’s DNA,” Morones-Ramírez explains. When the engineered bacteria replicates, the genetic circuit doesn’t become part of its genome.
Highly intelligent by bacterial standards, some multidrug resistant V. cholerae strains can also “collaborate” with other intestinal bacterial species to gain advantage and take hold of the gut.
In 2000, Boston-based researchers constructed an E.coli with a genetic switch that toggled between turning genes on and off two. Later, they built some safety checks into their bacteria. “To prevent unintentional or deleterious consequences, in 2009, we built a safety switch in the engineered bacteria’s genetic circuit that gets triggered after it gets exposed to a pathogen," says James Collins, a professor of biological engineering at MIT and faculty member at Harvard University’s Wyss Institute. “After getting rid of the pathogen, the engineered bacteria is designed to switch off and leave the patient's body.”

Overuse and misuse of antibiotics causes resistant strains to evolve
Adobe Stock
Seek and destroy
As the field of synthetic biology developed, scientists began using engineered bacteria to tackle superbugs. They first focused on Vibrio cholerae, which in the 19th and 20th century caused cholera pandemics in India, China, the Middle East, Europe, and Americas. Like many other bacteria, V. cholerae communicate with each other via quorum sensing, a process in which the microorganisms release different signaling molecules, to convey messages to its brethren. Highly intelligent by bacterial standards, some multidrug resistant V. cholerae strains can also “collaborate” with other intestinal bacterial species to gain advantage and take hold of the gut. When untreated, cholera has a mortality rate of 25 to 50 percent and outbreaks frequently occur in developing countries, especially during floods and droughts.
Sometimes, however, V. cholerae makes mistakes. In 2008, researchers at Cornell University observed that when quorum sensing V. cholerae accidentally released high concentrations of a signaling molecule called CAI-1, it had a counterproductive effect—the pathogen couldn’t colonize the gut.
So the group, led by John March, professor of biological and environmental engineering, developed a novel strategy to combat V. cholerae. They genetically engineered E.coli to eavesdrop on V. cholerae communication networks and equipped it with the ability to release the CAI-1 molecules. That interfered with V. cholerae progress. Two years later, the Cornell team showed that V. cholerae-infected mice treated with engineered E.coli had a 92 percent survival rate.
These findings inspired researchers to sic the good bacteria present in foods like yogurt and kimchi onto the drug-resistant ones.
Three years later in 2011, Singapore-based scientists engineered E.coli to detect and destroy Pseudomonas aeruginosa, an often drug-resistant pathogen that causes pneumonia, urinary tract infections, and sepsis. Once the genetically engineered E.coli found its target through its quorum sensing molecules, it then released a peptide, that could eradicate 99 percent of P. aeruginosa cells in a test-tube experiment. The team outlined their work in a Molecular Systems Biology study.
“At the time, we knew that we were entering new, uncharted territory,” says lead author Matthew Chang, an associate professor and synthetic biologist at the National University of Singapore and lead author of the study. “To date, we are still in the process of trying to understand how long these microbes stay in our bodies and how they might continue to evolve.”
More teams followed the same path. In a 2013 study, MIT researchers also genetically engineered E.coli to detect P. aeruginosa via the pathogen’s quorum-sensing molecules. It then destroyed the pathogen by secreting a lab-made toxin.
Probiotics that fight
A year later in 2014, a Nature study found that the abundance of Ruminococcus obeum, a probiotic bacteria naturally occurring in the human microbiome, interrupts and reduces V.cholerae’s colonization— by detecting the pathogen’s quorum sensing molecules. The natural accumulation of R. obeum in Bangladeshi adults helped them recover from cholera despite living in an area with frequent outbreaks.
The findings from 2008 to 2014 inspired Collins and his team to delve into how good bacteria present in foods like yogurt and kimchi can attack drug-resistant bacteria. In 2018, Collins and his team developed the engineered probiotic strategy. They tweaked a bacteria commonly found in yogurt called Lactococcus lactis to treat cholera.
Engineered bacteria can be trained to target pathogens when they are at their most vulnerable metabolic stage in the human gut. --José Rubén Morones-Ramírez.
More scientists followed with more experiments. So far, researchers have engineered various probiotic organisms to fight pathogenic bacteria like Staphylococcus aureus (leading cause of skin, tissue, bone, joint and blood infections) and Clostridium perfringens (which causes watery diarrhea) in test-tube and animal experiments. In 2020, Russian scientists engineered a probiotic called Pichia pastoris to produce an enzyme called lysostaphin that eradicated S. aureus in vitro. Another 2020 study from China used an engineered probiotic bacteria Lactobacilli casei as a vaccine to prevent C. perfringens infection in rabbits.
In a study last year, Ramírez’s group at the Autonomous University of Nuevo León, engineered E. coli to detect quorum-sensing molecules from Methicillin-resistant Staphylococcus aureus or MRSA, a notorious superbug. The E. coli then releases a bacteriocin that kills MRSA. “An antibiotic is just a molecule that is not intelligent,” says Ramírez. “On the other hand, engineered bacteria can be trained to target pathogens when they are at their most vulnerable metabolic stage in the human gut.”
Collins and Timothy Lu, an associate professor of biological engineering at MIT, found that engineered E. coli can help treat other conditions—such as phenylketonuria, a rare metabolic disorder, that causes the build-up of an amino acid phenylalanine. Their start-up Synlogic aims to commercialize the technology, and has completed a phase 2 clinical trial.
Circumventing the challenges
The bacteria-engineering technique is not without pitfalls. One major challenge is that beneficial gut bacteria produce their own quorum-sensing molecules that can be similar to those that pathogens secrete. If an engineered bacteria’s biosensor is not specific enough, it will be ineffective.
Another concern is whether engineered bacteria might mutate after entering the gut. “As with any technology, there are risks where bad actors could have the capability to engineer a microbe to act quite nastily,” says Collins of MIT. But Collins and Ramírez both insist that the chances of the engineered bacteria mutating on its own are virtually non-existent. “It is extremely unlikely for the engineered bacteria to mutate,” Ramírez says. “Coaxing a living cell to do anything on command is immensely challenging. Usually, the greater risk is that the engineered bacteria entirely lose its functionality.”
However, the biggest challenge is bringing the curative bacteria to consumers. Pharmaceutical companies aren’t interested in antibiotics or their alternatives because it’s less profitable than developing new medicines for non-infectious diseases. Unlike the more chronic conditions like diabetes or cancer that require long-term medications, infectious diseases are usually treated much quicker. Running clinical trials are expensive and antibiotic-alternatives aren’t lucrative enough.
“Unfortunately, new medications for antibiotic resistant infections have been pushed to the bottom of the field,” says Lu of MIT. “It's not because the technology does not work. This is more of a market issue. Because clinical trials cost hundreds of millions of dollars, the only solution is that governments will need to fund them.” Lu stresses that societies must lobby to change how the modern healthcare industry works. “The whole world needs better treatments for antibiotic resistance.”
Can blockchain help solve the Henrietta Lacks problem?
Marielle Gross, a professor at the University of Pittsburgh, shows patients a new app that tracks how their samples are used during biomedical research.
Science has come a long way since Henrietta Lacks, a Black woman from Baltimore, succumbed to cervical cancer at age 31 in 1951 -- only eight months after her diagnosis. Since then, research involving her cancer cells has advanced scientific understanding of the human papilloma virus, polio vaccines, medications for HIV/AIDS and in vitro fertilization.
Today, the World Health Organization reports that those cells are essential in mounting a COVID-19 response. But they were commercialized without the awareness or permission of Lacks or her family, who have filed a lawsuit against a biotech company for profiting from these “HeLa” cells.
While obtaining an individual's informed consent has become standard procedure before the use of tissues in medical research, many patients still don’t know what happens to their samples. Now, a new phone-based app is aiming to change that.
Tissue donors can track what scientists do with their samples while safeguarding privacy, through a pilot program initiated in October by researchers at the Johns Hopkins Berman Institute of Bioethics and the University of Pittsburgh’s Institute for Precision Medicine. The program uses blockchain technology to offer patients this opportunity through the University of Pittsburgh's Breast Disease Research Repository, while assuring that their identities remain anonymous to investigators.
A blockchain is a digital, tamper-proof ledger of transactions duplicated and distributed across a computer system network. Whenever a transaction occurs with a patient’s sample, multiple stakeholders can track it while the owner’s identity remains encrypted. Special certificates called “nonfungible tokens,” or NFTs, represent patients’ unique samples on a trusted and widely used blockchain that reinforces transparency.
Blockchain could be used to notify people if cancer researchers discover that they have certain risk factors.
“Healthcare is very data rich, but control of that data often does not lie with the patient,” said Julius Bogdan, vice president of analytics for North America at the Healthcare Information and Management Systems Society (HIMSS), a Chicago-based global technology nonprofit. “NFTs allow for the encapsulation of a patient’s data in a digital asset controlled by the patient.” He added that this technology enables a more secure and informed method of participating in clinical and research trials.
Without this technology, de-identification of patients’ samples during biomedical research had the unintended consequence of preventing them from discovering what researchers find -- even if that data could benefit their health. A solution was urgently needed, said Marielle Gross, assistant professor of obstetrics, gynecology and reproductive science and bioethics at the University of Pittsburgh School of Medicine.
“A researcher can learn something from your bio samples or medical records that could be life-saving information for you, and they have no way to let you or your doctor know,” said Gross, who is also an affiliate assistant professor at the Berman Institute. “There’s no good reason for that to stay the way that it is.”
For instance, blockchain could be used to notify people if cancer researchers discover that they have certain risk factors. Gross estimated that less than half of breast cancer patients are tested for mutations in BRCA1 and BRCA2 — tumor suppressor genes that are important in combating cancer. With normal function, these genes help prevent breast, ovarian and other cells from proliferating in an uncontrolled manner. If researchers find mutations, it’s relevant for a patient’s and family’s follow-up care — and that’s a prime example of how this newly designed app could play a life-saving role, she said.
Liz Burton was one of the first patients at the University of Pittsburgh to opt for the app -- called de-bi, which is short for decentralized biobank -- before undergoing a mastectomy for early-stage breast cancer in November, after it was diagnosed on a routine mammogram. She often takes part in medical research and looks forward to tracking her tissues.
“Anytime there’s a scientific experiment or study, I’m quick to participate -- to advance my own wellness as well as knowledge in general,” said Burton, 49, a life insurance service representative who lives in Carnegie, Pa. “It’s my way of contributing.”

Liz Burton was one of the first patients at the University of Pittsburgh to opt for the app before undergoing a mastectomy for early-stage breast cancer.
Liz Burton
The pilot program raises the issue of what investigators may owe study participants, especially since certain populations, such as Black and indigenous peoples, historically were not treated in an ethical manner for scientific purposes. “It’s a truly laudable effort,” Tamar Schiff, a postdoctoral fellow in medical ethics at New York University’s Grossman School of Medicine, said of the endeavor. “Research participants are beautifully altruistic.”
Lauren Sankary, a bioethicist and associate director of the neuroethics program at Cleveland Clinic, agrees that the pilot program provides increased transparency for study participants regarding how scientists use their tissues while acknowledging individuals’ contributions to research.
However, she added, “it may require researchers to develop a process for ongoing communication to be responsive to additional input from research participants.”
Peter H. Schwartz, professor of medicine and director of Indiana University’s Center for Bioethics in Indianapolis, said the program is promising, but he wonders what will happen if a patient has concerns about a particular research project involving their tissues.
“I can imagine a situation where a patient objects to their sample being used for some disease they’ve never heard about, or which carries some kind of stigma like a mental illness,” Schwartz said, noting that researchers would have to evaluate how to react. “There’s no simple answer to those questions, but the technology has to be assessed with an eye to the problems it could raise.”
To truly make a difference, blockchain must enable broad consent from patients, not just de-identification.
As a result, researchers may need to factor in how much information to share with patients and how to explain it, Schiff said. There are also concerns that in tracking their samples, patients could tell others what they learned before researchers are ready to publicly release this information. However, Bogdan, the vice president of the HIMSS nonprofit, believes only a minimal study identifier would be stored in an NFT, not patient data, research results or any type of proprietary trial information.
Some patients may be confused by blockchain and reluctant to embrace it. “The complexity of NFTs may prevent the average citizen from capitalizing on their potential or vendors willing to participate in the blockchain network,” Bogdan said. “Blockchain technology is also quite costly in terms of computational power and energy consumption, contributing to greenhouse gas emissions and climate change.”
In addition, this nascent, groundbreaking technology is immature and vulnerable to data security flaws, disputes over intellectual property rights and privacy issues, though it does offer baseline protections to maintain confidentiality. To truly make a difference, blockchain must enable broad consent from patients, not just de-identification, said Robyn Shapiro, a bioethicist and founding attorney at Health Sciences Law Group near Milwaukee.
The Henrietta Lacks story is a prime example, Shapiro noted. During her treatment for cervical cancer at Johns Hopkins, Lacks’s tissue was de-identified (albeit not entirely, because her cell line, HeLa, bore her initials). After her death, those cells were replicated and distributed for important and lucrative research and product development purposes without her knowledge or consent.
Nonetheless, Shapiro thinks that the initiative by the University of Pittsburgh and Johns Hopkins has potential to solve some ethical challenges involved in research use of biospecimens. “Compared to the system that allowed Lacks’s cells to be used without her permission, Shapiro said, “blockchain technology using nonfungible tokens that allow patients to follow their samples may enhance transparency, accountability and respect for persons who contribute their tissue and clinical data for research.”
Read more about laws that have prevented people from the rights to their own cells.
New tech for prison reform spreads to 11 states
The U.S. has the highest incarceration rate in the world, costing $182 billion per year, partly because its antiquated data systems often fail to identify people who should be released. A tech nonprofit is trying to change that.
A new non-profit called Recidiviz is using data technology to reduce the size of the U.S. criminal justice system. The bi-coastal company (SF and NYC) is currently working with 11 states to improve their systems and, so far, has helped remove nearly 69,000 people — ones left floundering in jail or on parole when they should have been released.
“The root cause is fragmentation,” says Clementine Jacoby, 31, a software engineer who worked at Google before co-founding Recidiviz in 2019. In the 1970s and 80s, the U.S. built a series of disconnected data systems, and this patchwork is still being used by criminal justice authorities today. It requires parole officers to manually calculate release dates, leading to errors in many cases. “[They] have done everything they need to do to earn their release, but they're still stuck in the system,” Jacoby says.
Recidiviz has built a platform that connects the different databases, with the goal of identifying people who are already qualified for release but remain behind bars or on supervision. “Think of Recidiviz like Google Maps,” says Jacoby, who worked on Maps when she was at the tech giant. Google Maps takes in data from different sources – satellite images, street maps, local business data — and organizes it into one easy view. “Recidiviz does something similar with criminal justice data,” Jacoby explains, “making it easy to identify people eligible to come home or to move to less intensive levels of supervision.”
People like Jacoby’s uncle. His experience with incarceration is what inspired her passion for criminal justice reform in the first place.
The problems are vast
The U.S. has the highest incarceration rate in the world — 2 million people according to the watchdog group, Prison Policy Initiative — at a cost of $182 billion a year. The numbers could be a lot lower if not for an array of problems including inaccurate sentencing calculations, flawed algorithms and parole violations laws.
Sentencing miscalculations
To determine eligibility for release, the current system requires corrections officers to check 21 different requirements spread across five different databases for each of the 90 to 100 people under their supervision. These manual calculations are time prohibitive, says Jacoby, and fall victim to human error.
In addition, Recidiviz found that policies aimed at helping to reduce the prison population don’t always work correctly. A key example is time off for good behavior laws that allow inmates to earn one day off for every 30 days of good behavior. Some states' data systems are built to calculate time off as one day per month of good behavior, rather than per day. Over the course of a decade-long sentence, Jacoby says these miscalculations can lead to a huge discrepancy in the calculated release data and the actual release date.
Algorithms
Commercial algorithm-based software systems for risk assessment continue to be widely used in the criminal justice system, even though a 2018 study published in Science Advances exposed their limitations. After the study went viral, it took three years for the Justice Department to issue a report on their own flawed algorithms used to reduce the federal prison population as part of the 2018 First Step Act. The program, it was determined, overestimated the risk of putting inmates of color into early-release programs.
Despite its name, Recidiviz does not build these types of algorithms for predicting recidivism, or whether someone will commit another crime after being released from prison. Rather, Jacoby says the company’s "descriptive analytics” approach is specifically intended to weed out incarceration inequalities and avoid algorithmic pitfalls.
Parole violation laws
Research shows that 350,000 people a year — about a quarter of the total prison population — are sent back not because they’ve committed another crime, but because they’ve broken a specific rule of their probation. “Things that wouldn't send you or I to prison, but would send someone on parole,” such as crossing county lines or being in the presence of alcohol when they shouldn’t be, are inflating the prison population, says Jacoby.
It’s personal for the co-founder and CEO
“I grew up with an uncle who went into the prison system,” Jacoby says. At 19, he was sentenced to ten years in prison for a non-violent crime. A few months after being released from jail, he was sent back for a non-violent parole violation.
“For my family, the fact that one in four prison admissions are driven not by a crime but by someone who's broken a rule on probation and parole was really profound because that happened to my uncle,” Jacoby says. The experience led her to begin studying criminal justice in high school, then college. She continued her dive into how the criminal justice system works as part of her Passion Project while at Google, a program that allows employees to spend 20 percent of their time on pro-bono work. Two colleagues whose family members had also been stuck in the system joined her.
As part of the project, Jacoby interviewed hundreds of people involved in the criminal justice system. “Those on the right, those on the left, agreed that bad data was slowing down reform,” she says. Their research brought them to North Dakota where they began to understand the root of the problem. The corrections department is making “huge, consequential decisions every day [without] … the data,” Jacoby says. In a new video by Recidiviz not yet released, Jacoby recounts her exchange with the state’s director of corrections who told her, “‘It’s not that we have the data and we just don’t know how to make it public; we don’t have the information you think we have.'"
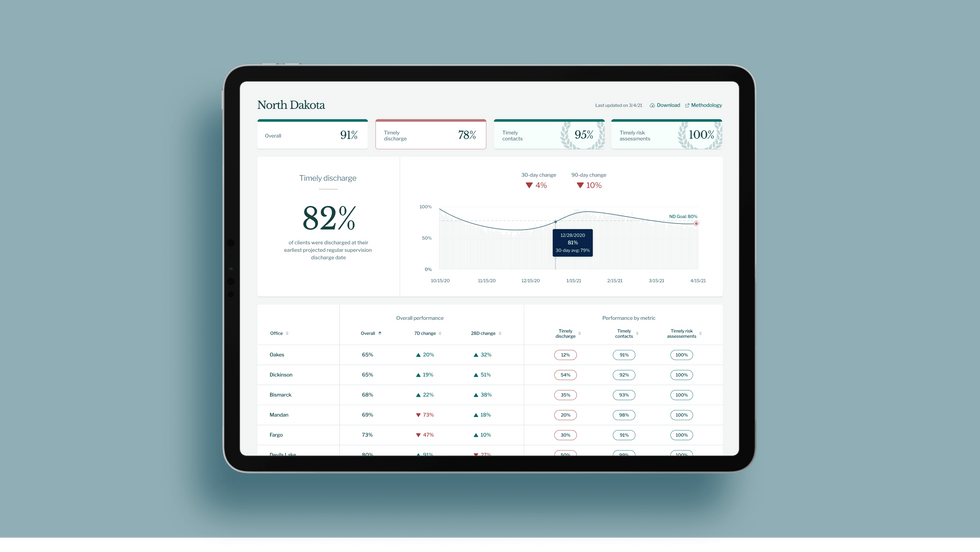
A mock-up (with fake data) of the types of dashboards and insights that Recidiviz provides to state governments.
Recidiviz
As a software engineer, Jacoby says the comment made no sense to her — until she witnessed it first-hand. “We spent a lot of time driving around in cars with corrections directors and parole officers watching them use these incredibly taxing, frankly terrible, old data systems,” Jacoby says.
As they weeded through thousands of files — some computerized, some on paper — they unearthed the consequences of bad data: Hundreds of people in prison well past their release date and thousands more whose release from parole was delayed because of minor paperwork issues. They found individuals stuck in parole because they hadn’t checked one last item off their eligibility list — like simply failing to provide their parole officer with a paystub. And, even when parolees advocated for themselves, the archaic system made it difficult for their parole officers to confirm their eligibility, so they remained in the system. Jacoby and her team also unpacked specific policies that drive racial disparities — such as fines and fees.
The Solution
It’s more than a trivial technical challenge to bring the incomplete, fragmented data onto a 21st century data platform. It takes months for Recidiviz to sift through a state’s information systems to connect databases “with the goal of tracking a person all the way through their journey and find out what’s working for 18- to 25-year-old men, what’s working for new mothers,” explains Jacoby in the video.
TED Talk: How bad data traps people in the U.S. justice system

TED Fellow Clementine Jacoby's TED Talk went live on Jan. 13. It describes how we can fix bad data in the criminal justice system, "bringing thousands of people home, reducing costs and improving public safety along the way."
Clementine Jacoby • TED2022
Ojmarrh Mitchell, an associate professor in the School of Criminology and Criminal Justice at Arizona State University, who is not involved with the company, says what Recidiviz is doing is “remarkable.” His perspective goes beyond academic analysis. In his pre-academic years, Mitchell was a probation officer, working within the framework of the “well known, but invisible” information sharing issues that plague criminal justice departments. The flexibility of Recidiviz’s approach is what makes it especially innovative, he says. “They identify the specific gaps in each jurisdiction and tailor a solution for that jurisdiction.”
On the downside, the process used by Recidiviz is “a bit opaque,” Mitchell says, with few details available on how Recidiviz designs its tools and tracks outcomes. By sharing more information about how its actions lead to progress in a given jurisdiction, Recidiviz could help reformers in other places figure out which programs have the best potential to work well.
The eleven states in which Recidiviz is working include California, Colorado, Maine, Michigan, Missouri, Pennsylvania and Tennessee. And a pilot program launched last year in Idaho, if scaled nationally, with could reduce the number of people in the criminal justice system by a quarter of a million people, Jacoby says. As part of the pilot, rather than relying on manual calculations, Recidiviz is equipping leaders and the probation officers with actionable information with a few clicks of an app that Recidiviz built.
Mitchell is disappointed that there’s even the need for Recidiviz. “This is a problem that government agencies have a responsibility to address,” he says. “But they haven’t.” For one company to come along and fill such a large gap is “remarkable.”

