Can You Trust Your Gut for Food Advice?
Kira Peikoff was the editor-in-chief of Leaps.org from 2017 to 2021. As a journalist, her work has appeared in The New York Times, Newsweek, Nautilus, Popular Mechanics, The New York Academy of Sciences, and other outlets. She is also the author of four suspense novels that explore controversial issues arising from scientific innovation: Living Proof, No Time to Die, Die Again Tomorrow, and Mother Knows Best. Peikoff holds a B.A. in Journalism from New York University and an M.S. in Bioethics from Columbia University. She lives in New Jersey with her husband and two young sons. Follow her on Twitter @KiraPeikoff.

Which foods are actually healthy for your individual gut microbiome? Several companies are offering personalized dietary guidance based on your test results, but their answers in one experiment turned up with some conflicting advice.
I recently got on the scale to weigh myself, thinking I've got to eat better. With so many trendy diets today claiming to improve health, from Keto to Paleo to Whole30, it can be confusing to figure out what we should and shouldn't eat for optimal nutrition.
A number of companies are now selling the concept of "personalized" nutrition based on the genetic makeup of your individual gut bugs.
My next thought was: I've got to lose a few pounds.
Consider a weird factoid: In addition to my fat, skin, bone and muscle, I'm carrying around two or three pounds of straight-up bacteria. Like you, I am the host to trillions of micro-organisms that live in my gut and are collectively known as my microbiome. An explosion of research has occurred in the last decade to try to understand exactly how these microbial populations, which are unique to each of us, may influence our overall health and potentially even our brains and behavior.
Lots of mysteries still remain, but it is established that these "bugs" are crucial to keeping our body running smoothly, performing functions like stimulating the immune system, synthesizing important vitamins, and aiding digestion. The field of microbiome science is evolving rapidly, and a number of companies are now selling the concept of "personalized" nutrition based on the genetic makeup of your individual gut bugs. The two leading players are Viome and DayTwo, but the landscape includes the newly launched startup Onegevity Health and others like Thryve, which offers customized probiotic supplements in addition to dietary recommendations.
The idea has immediate appeal – if science could tell you exactly what to make for lunch and what to avoid, you could forget about the fad diets and go with your own bespoke food pyramid. Wondering if the promise might be too good to be true, I decided to perform my own experiment.
Last fall, I sent the identical fecal sample to both Viome (I paid $425, but the price has since dropped to $299) and DayTwo ($349). A couple of months later, both reports finally arrived, and I eagerly opened each app to compare their recommendations.
First, I examined my results from Viome, which was founded in 2016 in Cupertino, Calif., and declares without irony on its website that "conflicting food advice is now obsolete."
I learned I have "average" metabolic fitness and "average" inflammatory activity in my gut, which are scores that the company defines based on a proprietary algorithm. But I have "low" microbial richness, with only 62 active species of bacteria identified in my sample, compared with the mean of 157 in their test population. I also received a list of the specific species in my gut, with names like Lactococcus and Romboutsia.
But none of it meant anything to me without actionable food advice, so I clicked through to the Recommendations page and found a list of My Superfoods (cranberry, garlic, kale, salmon, turmeric, watermelon, and bone broth) and My Foods to Avoid (chickpeas, kombucha, lentils, and rice noodles). There was also a searchable database of many foods that had been categorized for me, like "bell pepper; minimize" and "beef; enjoy."
"I just don't think sufficient data is yet available to make reliable personalized dietary recommendations based on one's microbiome."
Next, I looked at my results from DayTwo, which was founded in 2015 from research out of the Weizmann Institute of Science in Israel, and whose pitch to consumers is, "Blood sugar made easy. The algorithm diet personalized to you."
This app had some notable differences. There was no result about my metabolic fitness, microbial richness, or list of the species in my sample. There was also no list of superfoods or foods to avoid. Instead, the app encouraged me to build a meal by searching for foods in their database and combining them in beneficial ways for my blood sugar. Two slices of whole wheat bread received a score of 2.7 out of 10 ("Avoid"), but if combined with one cup of large curd cottage cheese, the score improved to 6.8 ("Limit"), and if I added two hard-boiled eggs, the score went up to 7.5 ("Good").
Perusing my list of foods with "Excellent" scores, I noticed some troubling conflicts with the other app. Lentils, which had been a no-no according to Viome, received high marks from DayTwo. Ditto for Kombucha. My purported superfood of cranberry received low marks. Almonds got an almost perfect score (9.7) while Viome told me to minimize them. I found similarly contradictory advice for foods I regularly eat, including navel oranges, peanuts, pork, and beets.
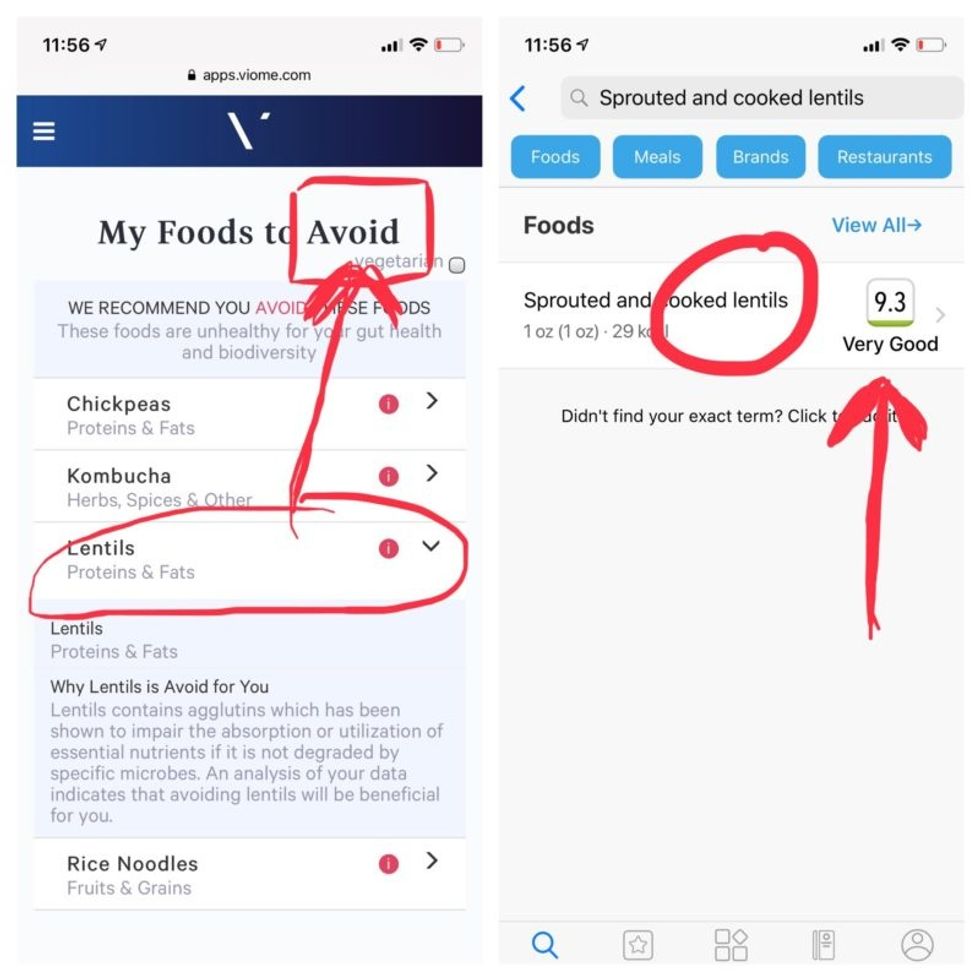
Contradictory dietary guidance that Kira Peikoff received from Viome (left) and DayTwo from an identical sample.
To be sure, there was some overlap. Both apps agreed on rice noodles (bad), chickpeas (bad), honey (bad), carrots (good), and avocado (good), among other foods.
But still, I was left scratching my head. Which set of recommendations should I trust, if either? And what did my results mean for the accuracy of this nascent field?
I called a couple of experts to find out.
"I have worked on the microbiome and nutrition for the last 20 years and I would be absolutely incapable of finding you evidence in the scientific literature that lentils have a detrimental effect based on the microbiome," said Dr. Jens Walter, an Associate Professor and chair for Nutrition, Microbes, and Gastrointestinal Health at the University of Alberta. "I just don't think sufficient data is yet available to make reliable personalized dietary recommendations based on one's microbiome. And even if they would have proprietary algorithms, at least one of them is not doing it right."
There is definite potential for personalized nutrition based on the microbiome, he said, but first, predictive models must be built and standardized, then linked to clinical endpoints, and tested in a large sample of healthy volunteers in order to enable extrapolations for the general population.
"It is mindboggling what you would need to do to make this work," he observed. "There are probably hundreds of relevant dietary compounds, then the microbiome has at least a hundred relevant species with a hundred or more relevant genes each, then you'd have to put all this together with relevant clinical outcomes. And there's a hundred-fold variation in that information between individuals."
However, Walter did acknowledge that the companies might be basing their algorithms on proprietary data that could potentially connect all the dots. I reached out to them to find out.
Amir Golan, the Chief Commercial Officer of DayTwo, told me, "It's important to emphasize this is a prediction, as the microbiome field is in a very early stage of research." But he added, "I believe we are the only company that has very solid science published in top journals and we can bring very actionable evidence and benefit to our uses."
He was referring to pioneering work out of the Weizmann Institute that was published in 2015 in the journal Cell, which logged the glycemic responses of 800 people in response to nearly 50,000 meals; adding information about the subjects' microbiomes enabled more accurate glycemic response predictions. Since then, Golan said, additional trials have been conducted, most recently with the Mayo Clinic, to duplicate the results, and other studies are ongoing whose results have not yet been published.
He also pointed out that the microbiome was merely one component that goes into building a client's profile, in addition to medical records, including blood glucose levels. (I provided my HbA1c levels, a measure of average blood sugar over the previous several months.)
"We are not saying we want to improve your gut microbiome. We provide a dynamic tool to help guide what you should eat to control your blood sugar and think about combinations," he said. "If you eat one thing, or with another, it will affect you in a different way."
Viome acknowledged that the two companies are taking very different approaches.
"DayTwo is primarily focused on the glycemic response," Naveen Jain, the CEO, told me. "If you can only eat butter for rest of your life, you will have no glycemic response but will probably die of a heart attack." He laughed. "Whereas we came from very different angle – what is happening inside the gut at a microbial level? When you eat food like spinach, how will that be metabolized in the gut? Will it produce the nutrients you need or cause inflammation?"
He said his team studied 1000 people who were on continuous glucose monitoring and fed them 45,000 meals, then built a proprietary data prediction model, looking at which microbes existed and how they actively broke down the food.
Jain pointed out that DayTwo sequences the DNA of the microbes, while Viome sequences the RNA – the active expression of DNA. That difference, in his opinion, is key to making accurate predictions.
"DNA is extremely stable, so when you eat any food and measure the DNA [in a fecal sample], you get all these false positives--you get DNA from plant food and meat, and you have no idea if those organisms are dead and simply transient, or actually exist. With RNA, you see what is actually alive in the gut."
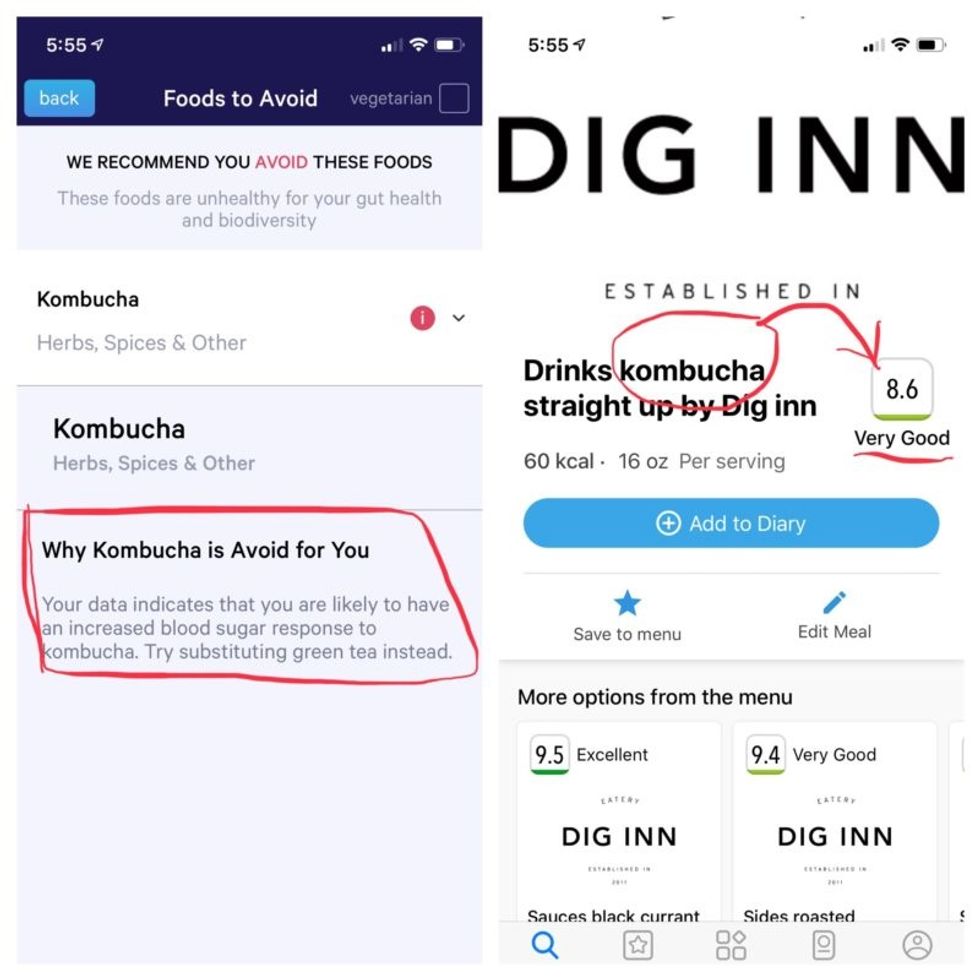
More contradictory food advice from Viome (left) and DayTwo.
Note that controversy exists over how it is possible with a fecal sample to effectively measure RNA, which degrades within minutes, though Jain said that his company has the technology to keep RNA stable for fourteen days.
Viome's approach, Jain maintains, is 90 percent accurate, based on as-yet unpublished data; a patent was filed just last week. DayTwo's approach is 66 percent accurate according to the latest published research.
Natasha Haskey, a registered dietician and doctoral student conducting research in the field of microbiome science and nutrition, is skeptical of both companies. "We can make broad statements, like eat more fruits and vegetables and fiber, but when it comes to specific foods, the science is just not there yet," she said. "I think there is a future, and we will be doing that someday, but not yet. Maybe we will be closer in ten years."
Professor Walter wholeheartedly agrees with Haskey, and suggested that if people want to eat a gut-healthy diet, they should focus on beneficial oils, fruits and vegetables, fish, a variety of whole grains, poultry and beans, and limit red meat and cheese, as well as avoid processed meats.
"These services are far over the tips of their science skis," Arthur Caplan, the founding head of New York University's Division of Medical Ethics, said in an email. "We simply don't know enough about the gut microbiome, its fluctuations and variability from person to person to support general [direct-to-consumer] testing. This is simply premature. We need standards for accuracy, specificity, and sensitivity, plus mandatory competent counseling for all such testing. They don't exist. Neither should DTC testing—yet."
Meanwhile, it's time for lunch. I close out my Viome and DayTwo apps and head to the kitchen to prepare a peanut butter sandwich. My gut tells me I'll be just fine.
Kira Peikoff was the editor-in-chief of Leaps.org from 2017 to 2021. As a journalist, her work has appeared in The New York Times, Newsweek, Nautilus, Popular Mechanics, The New York Academy of Sciences, and other outlets. She is also the author of four suspense novels that explore controversial issues arising from scientific innovation: Living Proof, No Time to Die, Die Again Tomorrow, and Mother Knows Best. Peikoff holds a B.A. in Journalism from New York University and an M.S. in Bioethics from Columbia University. She lives in New Jersey with her husband and two young sons. Follow her on Twitter @KiraPeikoff.
DNA- and RNA-based electronic implants may revolutionize healthcare
The test tubes contain tiny DNA/enzyme-based circuits, which comprise TRUMPET, a new type of electronic device, smaller than a cell.
Implantable electronic devices can significantly improve patients’ quality of life. A pacemaker can encourage the heart to beat more regularly. A neural implant, usually placed at the back of the skull, can help brain function and encourage higher neural activity. Current research on neural implants finds them helpful to patients with Parkinson’s disease, vision loss, hearing loss, and other nerve damage problems. Several of these implants, such as Elon Musk’s Neuralink, have already been approved by the FDA for human use.
Yet, pacemakers, neural implants, and other such electronic devices are not without problems. They require constant electricity, limited through batteries that need replacements. They also cause scarring. “The problem with doing this with electronics is that scar tissue forms,” explains Kate Adamala, an assistant professor of cell biology at the University of Minnesota Twin Cities. “Anytime you have something hard interacting with something soft [like muscle, skin, or tissue], the soft thing will scar. That's why there are no long-term neural implants right now.” To overcome these challenges, scientists are turning to biocomputing processes that use organic materials like DNA and RNA. Other promised benefits include “diagnostics and possibly therapeutic action, operating as nanorobots in living organisms,” writes Evgeny Katz, a professor of bioelectronics at Clarkson University, in his book DNA- And RNA-Based Computing Systems.
While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output.
Adamala’s research focuses on developing such biocomputing systems using DNA, RNA, proteins, and lipids. Using these molecules in the biocomputing systems allows the latter to be biocompatible with the human body, resulting in a natural healing process. In a recent Nature Communications study, Adamala and her team created a new biocomputing platform called TRUMPET (Transcriptional RNA Universal Multi-Purpose GatE PlaTform) which acts like a DNA-powered computer chip. “These biological systems can heal if you design them correctly,” adds Adamala. “So you can imagine a computer that will eventually heal itself.”
The basics of biocomputing
Biocomputing and regular computing have many similarities. Like regular computing, biocomputing works by running information through a series of gates, usually logic gates. A logic gate works as a fork in the road for an electronic circuit. The input will travel one way or another, giving two different outputs. An example logic gate is the AND gate, which has two inputs (A and B) and two different results. If both A and B are 1, the AND gate output will be 1. If only A is 1 and B is 0, the output will be 0 and vice versa. If both A and B are 0, the result will be 0. While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output. In this case, the DNA enters the logic gate as a single or double strand.
If the DNA is double-stranded, the system “digests” the DNA or destroys it, which results in non-fluorescence or “0” output. Conversely, if the DNA is single-stranded, it won’t be digested and instead will be copied by several enzymes in the biocomputing system, resulting in fluorescent RNA or a “1” output. And the output for this type of binary system can be expanded beyond fluorescence or not. For example, a “1” output might be the production of the enzyme insulin, while a “0” may be that no insulin is produced. “This kind of synergy between biology and computation is the essence of biocomputing,” says Stephanie Forrest, a professor and the director of the Biodesign Center for Biocomputing, Security and Society at Arizona State University.
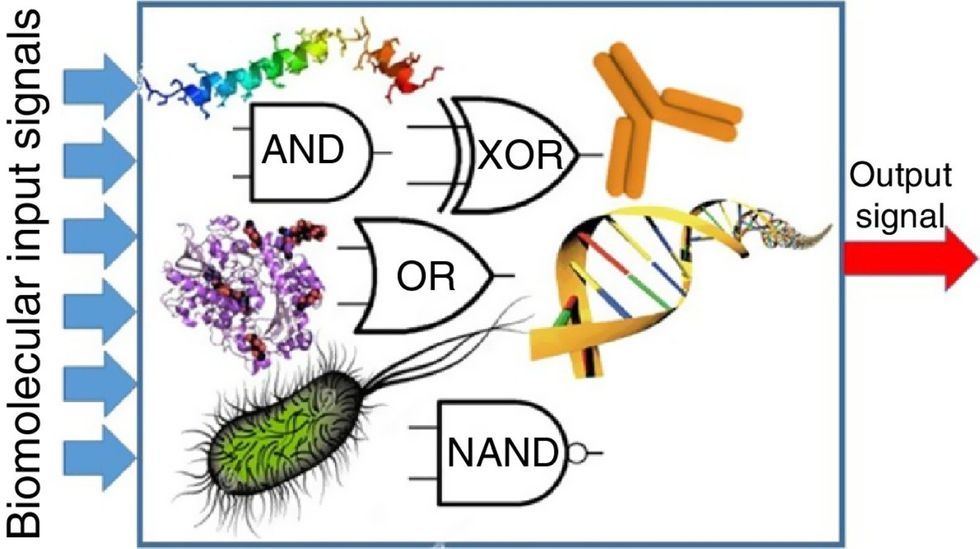
Biocomputing circles are made of DNA, RNA, proteins and even bacteria.
Evgeny Katz
The TRUMPET’s promise
Depending on whether the biocomputing system is placed directly inside a cell within the human body, or run in a test-tube, different environmental factors play a role. When an output is produced inside a cell, the cell's natural processes can amplify this output (for example, a specific protein or DNA strand), creating a solid signal. However, these cells can also be very leaky. “You want the cells to do the thing you ask them to do before they finish whatever their businesses, which is to grow, replicate, metabolize,” Adamala explains. “However, often the gate may be triggered without the right inputs, creating a false positive signal. So that's why natural logic gates are often leaky." While biocomputing outside a cell in a test tube can allow for tighter control over the logic gates, the outputs or signals cannot be amplified by a cell and are less potent.
TRUMPET, which is smaller than a cell, taps into both cellular and non-cellular biocomputing benefits. “At its core, it is a nonliving logic gate system,” Adamala states, “It's a DNA-based logic gate system. But because we use enzymes, and the readout is enzymatic [where an enzyme replicates the fluorescent RNA], we end up with signal amplification." This readout means that the output from the TRUMPET system, a fluorescent RNA strand, can be replicated by nearby enzymes in the platform, making the light signal stronger. "So it combines the best of both worlds,” Adamala adds.
These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body.
The TRUMPET biocomputing process is relatively straightforward. “If the DNA [input] shows up as single-stranded, it will not be digested [by the logic gate], and you get this nice fluorescent output as the RNA is made from the single-stranded DNA, and that's a 1,” Adamala explains. "And if the DNA input is double-stranded, it gets digested by the enzymes in the logic gate, and there is no RNA created from the DNA, so there is no fluorescence, and the output is 0." On the story's leading image above, if the tube is "lit" with a purple color, that is a binary 1 signal for computing. If it's "off" it is a 0.
While still in research, TRUMPET and other biocomputing systems promise significant benefits to personalized healthcare and medicine. These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body. The study’s lead author and graduate student Judee Sharon is already beginning to research TRUMPET's ability for earlier cancer diagnoses. Because the inputs for TRUMPET are single or double-stranded DNA, any mutated or cancerous DNA could theoretically be detected from the platform through the biocomputing process. Theoretically, devices like TRUMPET could be used to detect cancer and other diseases earlier.
Adamala sees TRUMPET not only as a detection system but also as a potential cancer drug delivery system. “Ideally, you would like the drug only to turn on when it senses the presence of a cancer cell. And that's how we use the logic gates, which work in response to inputs like cancerous DNA. Then the output can be the production of a small molecule or the release of a small molecule that can then go and kill what needs killing, in this case, a cancer cell. So we would like to develop applications that use this technology to control the logic gate response of a drug’s delivery to a cell.”
Although platforms like TRUMPET are making progress, a lot more work must be done before they can be used commercially. “The process of translating mechanisms and architecture from biology to computing and vice versa is still an art rather than a science,” says Forrest. “It requires deep computer science and biology knowledge,” she adds. “Some people have compared interdisciplinary science to fusion restaurants—not all combinations are successful, but when they are, the results are remarkable.”
Crickets are low on fat, high on protein, and can be farmed sustainably. They are also crunchy.
In today’s podcast episode, Leaps.org Deputy Editor Lina Zeldovich speaks about the health and ecological benefits of farming crickets for human consumption with Bicky Nguyen, who joins Lina from Vietnam. Bicky and her business partner Nam Dang operate an insect farm named CricketOne. Motivated by the idea of sustainable and healthy protein production, they started their unconventional endeavor a few years ago, despite numerous naysayers who didn’t believe that humans would ever consider munching on bugs.
Yet, making creepy crawlers part of our diet offers many health and planetary advantages. Food production needs to match the rise in global population, estimated to reach 10 billion by 2050. One challenge is that some of our current practices are inefficient, polluting and wasteful. According to nonprofit EarthSave.org, it takes 2,500 gallons of water, 12 pounds of grain, 35 pounds of topsoil and the energy equivalent of one gallon of gasoline to produce one pound of feedlot beef, although exact statistics vary between sources.
Meanwhile, insects are easy to grow, high on protein and low on fat. When roasted with salt, they make crunchy snacks. When chopped up, they transform into delicious pâtes, says Bicky, who invents her own cricket recipes and serves them at industry and public events. Maybe that’s why some research predicts that edible insects market may grow to almost $10 billion by 2030. Tune in for a delectable chat on this alternative and sustainable protein.
Listen on Apple | Listen on Spotify | Listen on Stitcher | Listen on Amazon | Listen on Google
Further reading:
More info on Bicky Nguyen
https://yseali.fulbright.edu.vn/en/faculty/bicky-n...
The environmental footprint of beef production
https://www.earthsave.org/environment.htm
https://www.watercalculator.org/news/articles/beef-king-big-water-footprints/
https://www.frontiersin.org/articles/10.3389/fsufs.2019.00005/full
https://ourworldindata.org/carbon-footprint-food-methane
Insect farming as a source of sustainable protein
https://www.insectgourmet.com/insect-farming-growing-bugs-for-protein/
https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/insect-farming
Cricket flour is taking the world by storm
https://www.cricketflours.com/
https://talk-commerce.com/blog/what-brands-use-cricket-flour-and-why/

Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.