Can You Trust Your Gut for Food Advice?
Kira Peikoff was the editor-in-chief of Leaps.org from 2017 to 2021. As a journalist, her work has appeared in The New York Times, Newsweek, Nautilus, Popular Mechanics, The New York Academy of Sciences, and other outlets. She is also the author of four suspense novels that explore controversial issues arising from scientific innovation: Living Proof, No Time to Die, Die Again Tomorrow, and Mother Knows Best. Peikoff holds a B.A. in Journalism from New York University and an M.S. in Bioethics from Columbia University. She lives in New Jersey with her husband and two young sons. Follow her on Twitter @KiraPeikoff.

Which foods are actually healthy for your individual gut microbiome? Several companies are offering personalized dietary guidance based on your test results, but their answers in one experiment turned up with some conflicting advice.
I recently got on the scale to weigh myself, thinking I've got to eat better. With so many trendy diets today claiming to improve health, from Keto to Paleo to Whole30, it can be confusing to figure out what we should and shouldn't eat for optimal nutrition.
A number of companies are now selling the concept of "personalized" nutrition based on the genetic makeup of your individual gut bugs.
My next thought was: I've got to lose a few pounds.
Consider a weird factoid: In addition to my fat, skin, bone and muscle, I'm carrying around two or three pounds of straight-up bacteria. Like you, I am the host to trillions of micro-organisms that live in my gut and are collectively known as my microbiome. An explosion of research has occurred in the last decade to try to understand exactly how these microbial populations, which are unique to each of us, may influence our overall health and potentially even our brains and behavior.
Lots of mysteries still remain, but it is established that these "bugs" are crucial to keeping our body running smoothly, performing functions like stimulating the immune system, synthesizing important vitamins, and aiding digestion. The field of microbiome science is evolving rapidly, and a number of companies are now selling the concept of "personalized" nutrition based on the genetic makeup of your individual gut bugs. The two leading players are Viome and DayTwo, but the landscape includes the newly launched startup Onegevity Health and others like Thryve, which offers customized probiotic supplements in addition to dietary recommendations.
The idea has immediate appeal – if science could tell you exactly what to make for lunch and what to avoid, you could forget about the fad diets and go with your own bespoke food pyramid. Wondering if the promise might be too good to be true, I decided to perform my own experiment.
Last fall, I sent the identical fecal sample to both Viome (I paid $425, but the price has since dropped to $299) and DayTwo ($349). A couple of months later, both reports finally arrived, and I eagerly opened each app to compare their recommendations.
First, I examined my results from Viome, which was founded in 2016 in Cupertino, Calif., and declares without irony on its website that "conflicting food advice is now obsolete."
I learned I have "average" metabolic fitness and "average" inflammatory activity in my gut, which are scores that the company defines based on a proprietary algorithm. But I have "low" microbial richness, with only 62 active species of bacteria identified in my sample, compared with the mean of 157 in their test population. I also received a list of the specific species in my gut, with names like Lactococcus and Romboutsia.
But none of it meant anything to me without actionable food advice, so I clicked through to the Recommendations page and found a list of My Superfoods (cranberry, garlic, kale, salmon, turmeric, watermelon, and bone broth) and My Foods to Avoid (chickpeas, kombucha, lentils, and rice noodles). There was also a searchable database of many foods that had been categorized for me, like "bell pepper; minimize" and "beef; enjoy."
"I just don't think sufficient data is yet available to make reliable personalized dietary recommendations based on one's microbiome."
Next, I looked at my results from DayTwo, which was founded in 2015 from research out of the Weizmann Institute of Science in Israel, and whose pitch to consumers is, "Blood sugar made easy. The algorithm diet personalized to you."
This app had some notable differences. There was no result about my metabolic fitness, microbial richness, or list of the species in my sample. There was also no list of superfoods or foods to avoid. Instead, the app encouraged me to build a meal by searching for foods in their database and combining them in beneficial ways for my blood sugar. Two slices of whole wheat bread received a score of 2.7 out of 10 ("Avoid"), but if combined with one cup of large curd cottage cheese, the score improved to 6.8 ("Limit"), and if I added two hard-boiled eggs, the score went up to 7.5 ("Good").
Perusing my list of foods with "Excellent" scores, I noticed some troubling conflicts with the other app. Lentils, which had been a no-no according to Viome, received high marks from DayTwo. Ditto for Kombucha. My purported superfood of cranberry received low marks. Almonds got an almost perfect score (9.7) while Viome told me to minimize them. I found similarly contradictory advice for foods I regularly eat, including navel oranges, peanuts, pork, and beets.
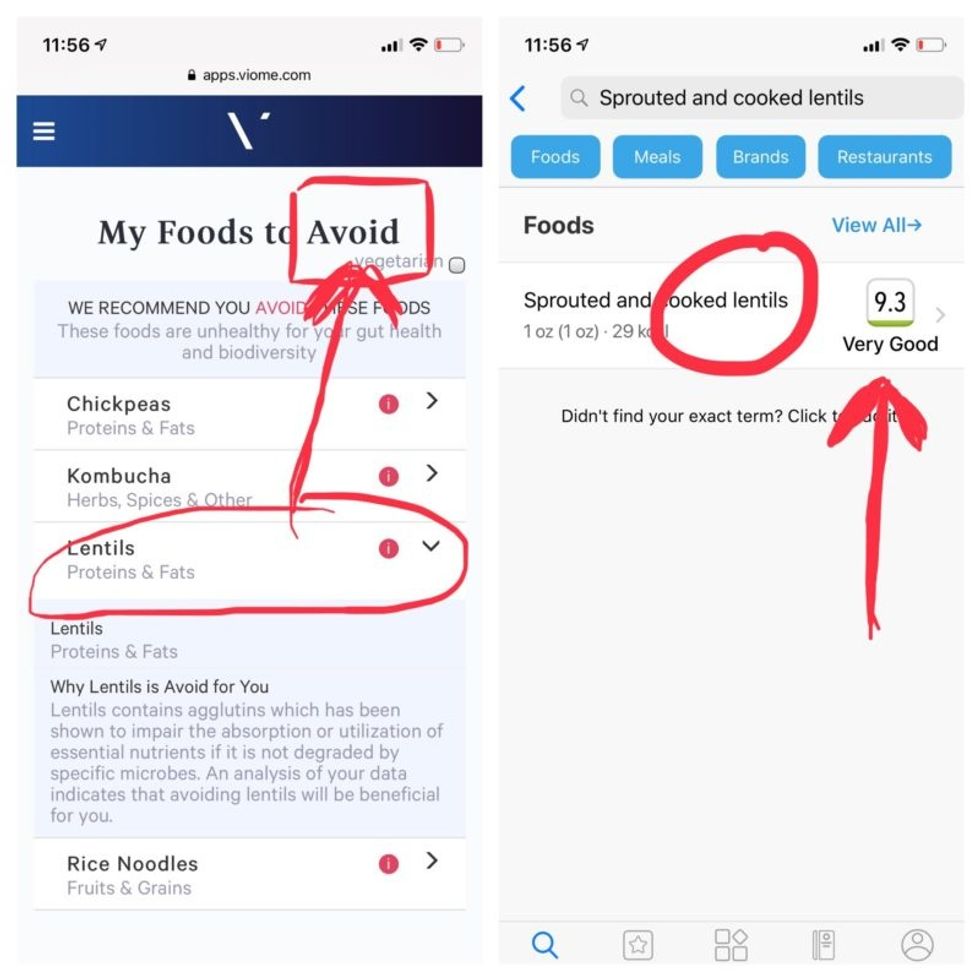
Contradictory dietary guidance that Kira Peikoff received from Viome (left) and DayTwo from an identical sample.
To be sure, there was some overlap. Both apps agreed on rice noodles (bad), chickpeas (bad), honey (bad), carrots (good), and avocado (good), among other foods.
But still, I was left scratching my head. Which set of recommendations should I trust, if either? And what did my results mean for the accuracy of this nascent field?
I called a couple of experts to find out.
"I have worked on the microbiome and nutrition for the last 20 years and I would be absolutely incapable of finding you evidence in the scientific literature that lentils have a detrimental effect based on the microbiome," said Dr. Jens Walter, an Associate Professor and chair for Nutrition, Microbes, and Gastrointestinal Health at the University of Alberta. "I just don't think sufficient data is yet available to make reliable personalized dietary recommendations based on one's microbiome. And even if they would have proprietary algorithms, at least one of them is not doing it right."
There is definite potential for personalized nutrition based on the microbiome, he said, but first, predictive models must be built and standardized, then linked to clinical endpoints, and tested in a large sample of healthy volunteers in order to enable extrapolations for the general population.
"It is mindboggling what you would need to do to make this work," he observed. "There are probably hundreds of relevant dietary compounds, then the microbiome has at least a hundred relevant species with a hundred or more relevant genes each, then you'd have to put all this together with relevant clinical outcomes. And there's a hundred-fold variation in that information between individuals."
However, Walter did acknowledge that the companies might be basing their algorithms on proprietary data that could potentially connect all the dots. I reached out to them to find out.
Amir Golan, the Chief Commercial Officer of DayTwo, told me, "It's important to emphasize this is a prediction, as the microbiome field is in a very early stage of research." But he added, "I believe we are the only company that has very solid science published in top journals and we can bring very actionable evidence and benefit to our uses."
He was referring to pioneering work out of the Weizmann Institute that was published in 2015 in the journal Cell, which logged the glycemic responses of 800 people in response to nearly 50,000 meals; adding information about the subjects' microbiomes enabled more accurate glycemic response predictions. Since then, Golan said, additional trials have been conducted, most recently with the Mayo Clinic, to duplicate the results, and other studies are ongoing whose results have not yet been published.
He also pointed out that the microbiome was merely one component that goes into building a client's profile, in addition to medical records, including blood glucose levels. (I provided my HbA1c levels, a measure of average blood sugar over the previous several months.)
"We are not saying we want to improve your gut microbiome. We provide a dynamic tool to help guide what you should eat to control your blood sugar and think about combinations," he said. "If you eat one thing, or with another, it will affect you in a different way."
Viome acknowledged that the two companies are taking very different approaches.
"DayTwo is primarily focused on the glycemic response," Naveen Jain, the CEO, told me. "If you can only eat butter for rest of your life, you will have no glycemic response but will probably die of a heart attack." He laughed. "Whereas we came from very different angle – what is happening inside the gut at a microbial level? When you eat food like spinach, how will that be metabolized in the gut? Will it produce the nutrients you need or cause inflammation?"
He said his team studied 1000 people who were on continuous glucose monitoring and fed them 45,000 meals, then built a proprietary data prediction model, looking at which microbes existed and how they actively broke down the food.
Jain pointed out that DayTwo sequences the DNA of the microbes, while Viome sequences the RNA – the active expression of DNA. That difference, in his opinion, is key to making accurate predictions.
"DNA is extremely stable, so when you eat any food and measure the DNA [in a fecal sample], you get all these false positives--you get DNA from plant food and meat, and you have no idea if those organisms are dead and simply transient, or actually exist. With RNA, you see what is actually alive in the gut."
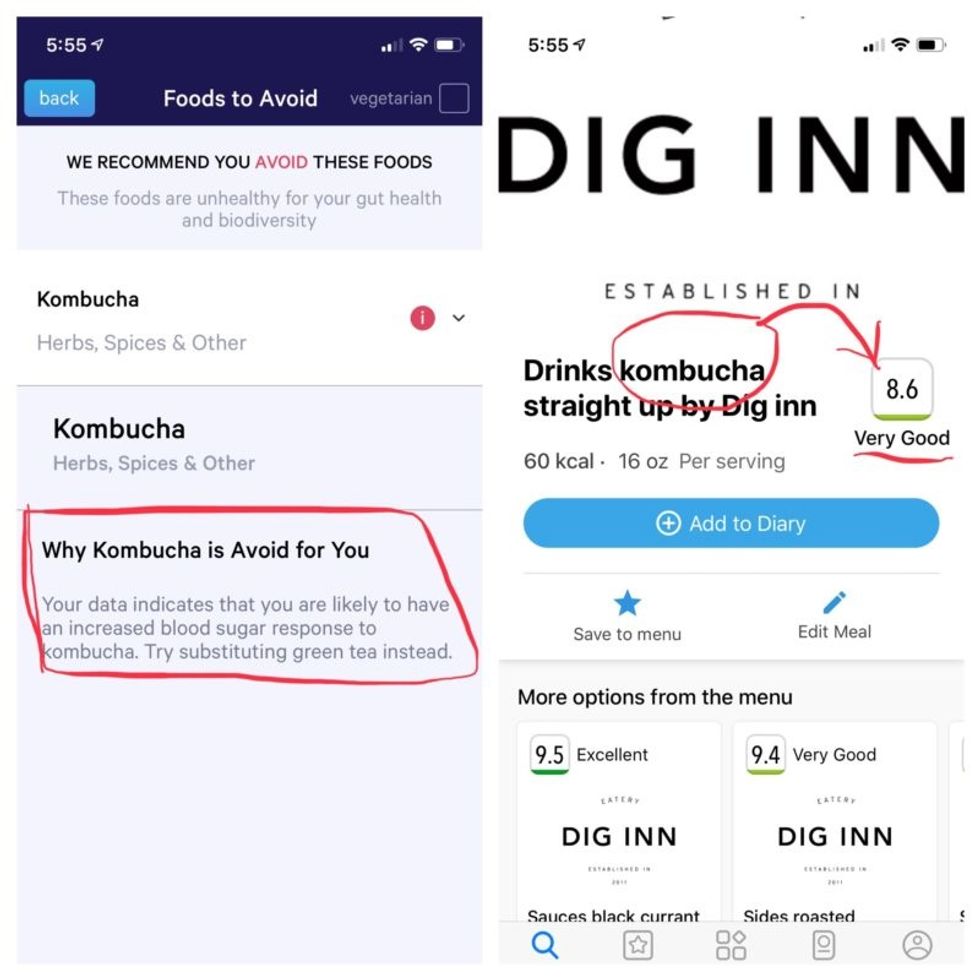
More contradictory food advice from Viome (left) and DayTwo.
Note that controversy exists over how it is possible with a fecal sample to effectively measure RNA, which degrades within minutes, though Jain said that his company has the technology to keep RNA stable for fourteen days.
Viome's approach, Jain maintains, is 90 percent accurate, based on as-yet unpublished data; a patent was filed just last week. DayTwo's approach is 66 percent accurate according to the latest published research.
Natasha Haskey, a registered dietician and doctoral student conducting research in the field of microbiome science and nutrition, is skeptical of both companies. "We can make broad statements, like eat more fruits and vegetables and fiber, but when it comes to specific foods, the science is just not there yet," she said. "I think there is a future, and we will be doing that someday, but not yet. Maybe we will be closer in ten years."
Professor Walter wholeheartedly agrees with Haskey, and suggested that if people want to eat a gut-healthy diet, they should focus on beneficial oils, fruits and vegetables, fish, a variety of whole grains, poultry and beans, and limit red meat and cheese, as well as avoid processed meats.
"These services are far over the tips of their science skis," Arthur Caplan, the founding head of New York University's Division of Medical Ethics, said in an email. "We simply don't know enough about the gut microbiome, its fluctuations and variability from person to person to support general [direct-to-consumer] testing. This is simply premature. We need standards for accuracy, specificity, and sensitivity, plus mandatory competent counseling for all such testing. They don't exist. Neither should DTC testing—yet."
Meanwhile, it's time for lunch. I close out my Viome and DayTwo apps and head to the kitchen to prepare a peanut butter sandwich. My gut tells me I'll be just fine.
Kira Peikoff was the editor-in-chief of Leaps.org from 2017 to 2021. As a journalist, her work has appeared in The New York Times, Newsweek, Nautilus, Popular Mechanics, The New York Academy of Sciences, and other outlets. She is also the author of four suspense novels that explore controversial issues arising from scientific innovation: Living Proof, No Time to Die, Die Again Tomorrow, and Mother Knows Best. Peikoff holds a B.A. in Journalism from New York University and an M.S. in Bioethics from Columbia University. She lives in New Jersey with her husband and two young sons. Follow her on Twitter @KiraPeikoff.
Have You Heard of the Best Sport for Brain Health?
In this week's Friday Five, research points to this brain healthiest of sports. Plus, the natural way to reprogram cells to a younger state, the network that could underlie many different mental illnesses, and a new test could diagnose autism in newborns. Plus, scientists 3D print an ear and attach it to woman
The Friday Five covers five stories in research that you may have missed this week. There are plenty of controversies and troubling ethical issues in science – and we get into many of them in our online magazine – but this news roundup focuses on scientific creativity and progress to give you a therapeutic dose of inspiration headed into the weekend.
Listen on Apple | Listen on Spotify | Listen on Stitcher | Listen on Amazon | Listen on Google
Here are the promising studies covered in this week's Friday Five:
- Reprogram cells to a younger state
- Pick up this sport for brain health
- Do all mental illnesses have the same underlying cause?
- New test could diagnose autism in newborns
- Scientists 3D print an ear and attach it to woman
Can blockchain help solve the Henrietta Lacks problem?
Marielle Gross, a professor at the University of Pittsburgh, shows patients a new app that tracks how their samples are used during biomedical research.
Science has come a long way since Henrietta Lacks, a Black woman from Baltimore, succumbed to cervical cancer at age 31 in 1951 -- only eight months after her diagnosis. Since then, research involving her cancer cells has advanced scientific understanding of the human papilloma virus, polio vaccines, medications for HIV/AIDS and in vitro fertilization.
Today, the World Health Organization reports that those cells are essential in mounting a COVID-19 response. But they were commercialized without the awareness or permission of Lacks or her family, who have filed a lawsuit against a biotech company for profiting from these “HeLa” cells.
While obtaining an individual's informed consent has become standard procedure before the use of tissues in medical research, many patients still don’t know what happens to their samples. Now, a new phone-based app is aiming to change that.
Tissue donors can track what scientists do with their samples while safeguarding privacy, through a pilot program initiated in October by researchers at the Johns Hopkins Berman Institute of Bioethics and the University of Pittsburgh’s Institute for Precision Medicine. The program uses blockchain technology to offer patients this opportunity through the University of Pittsburgh's Breast Disease Research Repository, while assuring that their identities remain anonymous to investigators.
A blockchain is a digital, tamper-proof ledger of transactions duplicated and distributed across a computer system network. Whenever a transaction occurs with a patient’s sample, multiple stakeholders can track it while the owner’s identity remains encrypted. Special certificates called “nonfungible tokens,” or NFTs, represent patients’ unique samples on a trusted and widely used blockchain that reinforces transparency.
Blockchain could be used to notify people if cancer researchers discover that they have certain risk factors.
“Healthcare is very data rich, but control of that data often does not lie with the patient,” said Julius Bogdan, vice president of analytics for North America at the Healthcare Information and Management Systems Society (HIMSS), a Chicago-based global technology nonprofit. “NFTs allow for the encapsulation of a patient’s data in a digital asset controlled by the patient.” He added that this technology enables a more secure and informed method of participating in clinical and research trials.
Without this technology, de-identification of patients’ samples during biomedical research had the unintended consequence of preventing them from discovering what researchers find -- even if that data could benefit their health. A solution was urgently needed, said Marielle Gross, assistant professor of obstetrics, gynecology and reproductive science and bioethics at the University of Pittsburgh School of Medicine.
“A researcher can learn something from your bio samples or medical records that could be life-saving information for you, and they have no way to let you or your doctor know,” said Gross, who is also an affiliate assistant professor at the Berman Institute. “There’s no good reason for that to stay the way that it is.”
For instance, blockchain could be used to notify people if cancer researchers discover that they have certain risk factors. Gross estimated that less than half of breast cancer patients are tested for mutations in BRCA1 and BRCA2 — tumor suppressor genes that are important in combating cancer. With normal function, these genes help prevent breast, ovarian and other cells from proliferating in an uncontrolled manner. If researchers find mutations, it’s relevant for a patient’s and family’s follow-up care — and that’s a prime example of how this newly designed app could play a life-saving role, she said.
Liz Burton was one of the first patients at the University of Pittsburgh to opt for the app -- called de-bi, which is short for decentralized biobank -- before undergoing a mastectomy for early-stage breast cancer in November, after it was diagnosed on a routine mammogram. She often takes part in medical research and looks forward to tracking her tissues.
“Anytime there’s a scientific experiment or study, I’m quick to participate -- to advance my own wellness as well as knowledge in general,” said Burton, 49, a life insurance service representative who lives in Carnegie, Pa. “It’s my way of contributing.”

Liz Burton was one of the first patients at the University of Pittsburgh to opt for the app before undergoing a mastectomy for early-stage breast cancer.
Liz Burton
The pilot program raises the issue of what investigators may owe study participants, especially since certain populations, such as Black and indigenous peoples, historically were not treated in an ethical manner for scientific purposes. “It’s a truly laudable effort,” Tamar Schiff, a postdoctoral fellow in medical ethics at New York University’s Grossman School of Medicine, said of the endeavor. “Research participants are beautifully altruistic.”
Lauren Sankary, a bioethicist and associate director of the neuroethics program at Cleveland Clinic, agrees that the pilot program provides increased transparency for study participants regarding how scientists use their tissues while acknowledging individuals’ contributions to research.
However, she added, “it may require researchers to develop a process for ongoing communication to be responsive to additional input from research participants.”
Peter H. Schwartz, professor of medicine and director of Indiana University’s Center for Bioethics in Indianapolis, said the program is promising, but he wonders what will happen if a patient has concerns about a particular research project involving their tissues.
“I can imagine a situation where a patient objects to their sample being used for some disease they’ve never heard about, or which carries some kind of stigma like a mental illness,” Schwartz said, noting that researchers would have to evaluate how to react. “There’s no simple answer to those questions, but the technology has to be assessed with an eye to the problems it could raise.”
To truly make a difference, blockchain must enable broad consent from patients, not just de-identification.
As a result, researchers may need to factor in how much information to share with patients and how to explain it, Schiff said. There are also concerns that in tracking their samples, patients could tell others what they learned before researchers are ready to publicly release this information. However, Bogdan, the vice president of the HIMSS nonprofit, believes only a minimal study identifier would be stored in an NFT, not patient data, research results or any type of proprietary trial information.
Some patients may be confused by blockchain and reluctant to embrace it. “The complexity of NFTs may prevent the average citizen from capitalizing on their potential or vendors willing to participate in the blockchain network,” Bogdan said. “Blockchain technology is also quite costly in terms of computational power and energy consumption, contributing to greenhouse gas emissions and climate change.”
In addition, this nascent, groundbreaking technology is immature and vulnerable to data security flaws, disputes over intellectual property rights and privacy issues, though it does offer baseline protections to maintain confidentiality. To truly make a difference, blockchain must enable broad consent from patients, not just de-identification, said Robyn Shapiro, a bioethicist and founding attorney at Health Sciences Law Group near Milwaukee.
The Henrietta Lacks story is a prime example, Shapiro noted. During her treatment for cervical cancer at Johns Hopkins, Lacks’s tissue was de-identified (albeit not entirely, because her cell line, HeLa, bore her initials). After her death, those cells were replicated and distributed for important and lucrative research and product development purposes without her knowledge or consent.
Nonetheless, Shapiro thinks that the initiative by the University of Pittsburgh and Johns Hopkins has potential to solve some ethical challenges involved in research use of biospecimens. “Compared to the system that allowed Lacks’s cells to be used without her permission, Shapiro said, “blockchain technology using nonfungible tokens that allow patients to follow their samples may enhance transparency, accountability and respect for persons who contribute their tissue and clinical data for research.”
Read more about laws that have prevented people from the rights to their own cells.

