COVID Variants Are Like “a Thief Changing Clothes” – and Our Camera System Barely Exists

Being able to track variants of concern in real time is crucial to our ability to stay ahead of the virus.
Whether it's "natural selection" as Darwin called it, or it's "mutating" as the X-Men called it, living organisms change over time, developing thumbs or more efficient protein spikes, depending on the organism and the demands of its environment. The coronavirus that causes COVID-19, SARS-CoV-2, is not an exception, and now, after the virus has infected millions of people around the globe for more than a year, scientists are beginning to see those changes.
The notorious variants that have popped up include B.1.1.7, sometimes called the UK variant, as well as P.1 and B.1.351, which seem to have emerged in Brazil and South Africa respectively. As vaccinations are picking up pace, officials are warning that now
is not the time to become complacent or relax restrictions because the variants aren't well understood.
Some appear to be more transmissible, and deadlier, while others can evade the immune system's defenses better than earlier versions of the virus, potentially undermining the effectiveness of vaccines to some degree. Genomic surveillance, the process of sequencing the genetic code of the virus widely to observe changes and patterns, is a critical way that scientists can keep track of its evolution and work to understand how the variants might affect humans.
"It's like a thief changing clothes"
It's important to note that viruses mutate all the time. If there were funding and personnel to sequence the genome of every sample of the virus, scientists would see thousands of mutations. Not every variant deserves our attention. The vast majority of mutations are not important at all, but recognizing those that are is a crucial tool in getting and staying ahead of the virus. The work of sequencing, analyzing, observing patterns, and using public health tools as necessary is complicated and confusing to those without years of specialized training.
Jeremy Kamil, associate professor of microbiology and immunology at LSU Health Shreveport, in Louisiana, says that the variants developing are like a thief changing clothes. The thief goes in your house, steals your stuff, then leaves and puts on a different shirt and a wig, in the hopes you won't recognize them. Genomic surveillance catches the "thief" even in those different clothes.
One of the tricky things about variants is recognizing the point at which they move from interesting, to concerning at a local level, to dangerous in a larger context.
Understanding variants, both the uninteresting ones and the potentially concerning ones, gives public health officials and researchers at different levels a useful set of tools. Locally, knowing which variants are circulating in the community helps leaders know whether mask mandates and similar measures should be implemented or discontinued, or whether businesses and schools can open relatively safely.
There's more to it than observing new variants
Analysis is complex, particularly when it comes to understanding which variants are of concern. "So the question is always if a mutation becomes common, is that a random occurrence?" says Phoebe Lostroh, associate professor of molecular biology at Colorado College. "Or is the variant the result of some kind of selection because the mutation changes some property about the virus that makes it reproduce more quickly than variants of the virus that don't have that mutation? For a virus, [mutations can affect outcomes like] how much it replicates inside a person's body, how much somebody breathes it out, whether the particles that somebody might breathe in get smaller and can lead to greater transmission."
Along with all of those factors, accurate and useful genomic surveillance requires an understanding of where variants are occurring, how they are related, and an examination of why they might be prevalent.
For example, if a potentially worrisome variant appears in a community and begins to spread very quickly, it's not time to raise a public health alarm until several important questions have been answered, such as whether the variant is spreading due to specific events, or if it's happening because the mutation has allowed the virus to infect people more efficiently. Kamil offered a hypothetical scenario to explain: Imagine that a member of a community became infected and the virus mutated. That person went to church and three more people were infected, but one of them went to a karaoke bar and while singing infected 100 other people. Examining the conditions under which the virus has spread is, therefore, an essential part of untangling whether a mutation itself made the virus more transmissible or if an infected person's behaviors contributed to a local outbreak.
One of the tricky things about variants is recognizing the point at which they move from interesting, to concerning at a local level, to dangerous in a larger context. Genomic sequencing can help with that, but only when it's coordinated. When the same mutation occurs frequently, but is localized to one region, it's a concern, but when the same mutation happens in different places at the same time, it's much more likely that the "virus is learning that's a good mutation," explains Kamil.
The process is called convergent evolution, and it was a fascinating topic long before COVID. Just as your heritage can be traced through DNA, so can that of viruses, and when separate lineages develop similar traits it's almost like scientists can see evolution happening in real time. A mutation to SARS-CoV-2 that happens in more than one place at once is a mutation that makes it easier in some way for the virus to survive and that is when it may become alarming. The widespread, documented variants P.1 and B.1.351 are examples of convergence because they share some of the same virulent mutations despite having developed thousands of miles apart.
However, even variants that are emerging in different places at the same time don't present the kind of threat SARS-CoV-2 did in 2019. "This is nature," says Kamil. "It just means that this virus will not easily be driven to extinction or complete elimination by vaccines." Although a person who has already had COVID-19 can be reinfected with a variant, "it is almost always much milder disease" than the original infection, Kamil adds. Rather than causing full-fledged disease, variants have the potiental to "penetrate herd immunity, spreading relatively quietly among people who have developed natural immunity or been vaccinated, until the virus finds someone who has no immunity yet, and that person would be at risk of hospitalization-grade severe disease or death."
Surveillance and predictions
According to Lostroh, genomic surveillance can help scientists predict what's going to happen. "With the British strain, for instance, that's more transmissible, you can measure how fast it's doubling in the population and you can sort of tell whether we should take more measures against this mutation. Should we shut things down a little longer because that mutation is present in the population? That could be really useful if you did enough sampling in the population that you knew where it was," says Lostroh. If, for example, the more transmissible strain was present in 50 percent of cases, but in another county or state it was barely present, it would allow for rolling lockdowns instead of sweeping measures.
Variants are also extremely important when it comes to the development, manufacture, and distribution of vaccines. "You're also looking at medical countermeasures, such as whether your vaccine is still effective, or if your antiviral needs to be updated," says Lane Warmbrod, a senior analyst and research associate at Johns Hopkins Center for Health Security.
Properly funded and extensive genomic surveillance could eventually help control endemic diseases, too, like the seasonal flu, or other common respiratory infections. Kamil says he envisions a future in which genomic surveillance allows for prediction of sickness just as the weather is predicted today. "It's a 51 for infection today at the San Francisco Airport. There's been detection of some respiratory viruses," he says, offering an example. He says that if you're a vulnerable person, if you're immune-suppressed for some reason, you may want to wear a mask based on the sickness report.
The U.S. has the ability, but lacks standards
The benefits of widespread genomic surveillance are clear, and the United States certainly has the necessary technology, equipment, and personnel to carry it out. But, it's not happening at the speed and extent it needs to for the country to gain the benefits.
"The numbers are improving," said Kamil. "We're probably still at less than half a percent of all the samples that have been taken have been sequenced since the beginning of the pandemic."
Although there's no consensus on how many sequences is ideal for a robust surveillance program, modeling performed by the company Illumina suggests about 5 percent of positive tests should be sequenced. The reasons the U.S. has lagged in implementing a sequencing program are complex and varied, but solvable.
Perhaps the most important element that is currently missing is leadership. In order to conduct an effective genomic surveillance program, there need to be standards. The Johns Hopkins Center for Health Security recently published a paper with recommendations as to what kinds of elements need to be standardized in order to make the best use of sequencing technology and analysis.
"Along with which bioinformatic pipelines you're going to use to do the analyses, which sequencing strategy protocol are you going to use, what's your sampling strategy going to be, how is the data is going to be reported, what data gets reported," says Warmbrod. Currently, there's no guidance from the CDC on any of those things. So, while scientists can collect and report information, they may be collecting and reporting different information that isn't comparable, making it less useful for public health measures and vaccine updates.
Globally, one of the most important tools in making the information from genomic surveillance useful is GISAID, a platform designed for scientists to share -- and, importantly, to be credited for -- their data regarding genetic sequences of influenza. Originally, it was launched as a database of bird flu sequences, but has evolved to become an essential tool used by the WHO to make flu vaccine virus recommendations each year. Scientists who share their credentials have free access to the database, and anyone who uses information from the database must credit the scientist who uploaded that information.
Safety, logistics, and funding matter
Scientists at university labs and other small organizations have been uploading sequences to GISAID almost from the beginning of the pandemic, but their funding is generally limited, and there are no standards regarding information collection or reporting. Private, for-profit labs haven't had motivation to set up sequencing programs, although many of them have the logistical capabilities and funding to do so. Public health departments are understaffed, underfunded, and overwhelmed.
University labs may also be limited by safety concerns. The SARS-CoV-2 virus is dangerous, and there's a question of how samples should be transported to labs for sequencing.
Larger, for-profit organizations often have the tools and distribution capabilities to safely collect and sequence samples, but there hasn't been a profit motive. Genomic sequencing is less expensive now than ever before, but even at $100 per sample, the cost adds up -- not to mention the cost of employing a scientist with the proper credentials to analyze the sequence.
The path forward
The recently passed COVID-19 relief bill does have some funding to address genomic sequencing. Specifically, the American Rescue Plan Act includes $1.75 billion in funding for the Centers for Disease Control and Prevention's Advanced Molecular Detection (AMD) program. In an interview last month, CDC Director Rochelle Walensky said that the additional funding will be "a dial. And we're going to need to dial it up." AMD has already announced a collaboration called the Sequencing for Public Health Emergency Response, Epidemiology, and Surveillance (SPHERES) Initiative that will bring together scientists from public health, academic, clinical, and non-profit laboratories across the country with the goal of accelerating sequencing.
Such a collaboration is a step toward following the recommendations in the paper Warmbrod coauthored. Building capacity now, creating a network of labs, and standardizing procedures will mean improved health in the future. "I want to be optimistic," she says. "The good news is there are a lot of passionate, smart, capable people who are continuing to work with government and work with different stakeholders." She cautions, however, that without a national strategy we won't succeed.
"If we maximize the potential and create that framework now, we can also use it for endemic diseases," she says. "It's a very helpful system for more than COVID if we're smart in how we plan it."
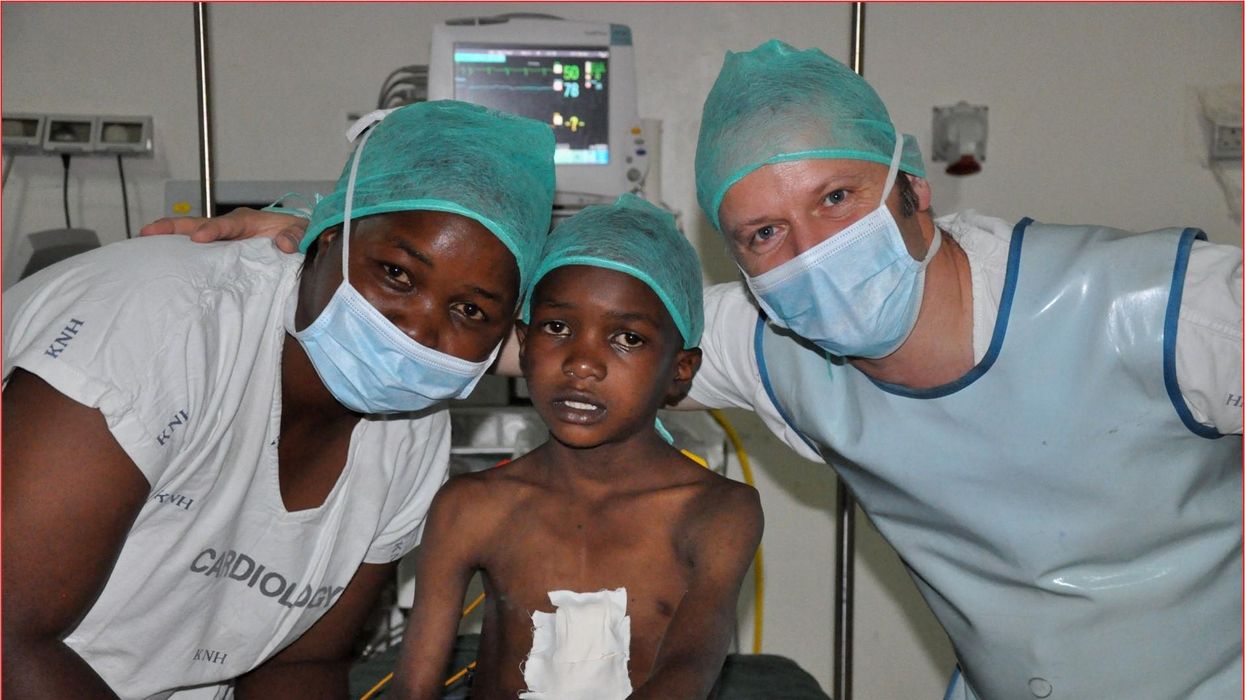
These doctors have a heart for recycling
In the U.S. and Europe, it is illegal to reuse pacemakers and other implants. Therefore, cardiologists export them to the global South where they save the lives of people of all ages.
This is part 3 of a three part series on a new generation of doctors leading the charge to make the health care industry more sustainable - for the benefit of their patients and the planet. Read part 1 here and part 2 here.
One could say that over 400 people owe their life to the fact that Carsten Israel fell in love. Twenty years ago, as a young doctor in Frankfurt, Germany, he began to court an au pair from Kenya, Elisabeth, his now-wife of 13 years with whom he has three children. When the couple started visiting her parents in Kenya, Israel wanted to check out the local hospitals, “just out of professional curiosity,“ says the cardiologist, who is currently the head doctor at the Clinic for Interior Medicine in Bielefeld. “I was completely shocked.“
Often he observed there were no doctors in the E.R.s, and hte nurses could render only basic first aid. “When somebody fell into a coma, they fell into a coma,“ Israel remembers. “There weren’t even any defibrillators to restart a patient’s heart,” while defibrillators are standard equipment in most clinics in the U.S. and Europe as lifesaving devices. When Israel finally visited the largest and most modern hospital in Nairobi, he found it better equipped but he learned that its services were only available to patients who could afford them. The cardiologist there had a drawer full of petitions from patients with heart ailments who couldn’t afford lifesaving surgery. Even two decades ago, a pacemaker cost $5,000 in Kenya, which made it unaffordable for most Kenyans who earn an average of $600 per month.
Since 2003, Israel and a team of two doctors and two nurses visit Kenya and Zambia once or twice a year to implant German pacemakers for free. Notably, the pacemakers and defibrillators Israel exports to Africa would end up in the landfill in Germany. Clinics have to pay for specialized services to dispose of this medical equipment. “In Germany, I could go to jail if I used a defibrillator that is one day past its expiration date,“ Israel says, “but in Kenya, people don’t have the money for the cheapest model. What nonsense to throw this precious medical equipment away while people in poorer countries die because they desperately need it.“
Israel works at the breakpoint between the laws in a wealthy country like Germany and the reality in the global South. The U.S. and most European countries have strict laws that ban the reuse of medical implants and enforce strict expiration dates for medical equipment. “But if a pacemaker is a few days past its expiration date, it still works perfectly fine,“ Israel says. “And it also happens that we implant a pacemaker and five months later it turns out that the patient needs a different kind. Then we replace it and we’d have to trash the first one in Germany, though it could easily run another 12 years.“
“If we get this right, we have lots of devices we can implant, hips and knees, etcetera. Where this will lead is limitless," says Eva Kline Rogers, the program coordinator for My Heart, Your Heart.
Israel has been collecting donations of pacemakers and defibrillators from manufacturers but also from other doctors and from funeral homes for his nonprofit Pacemakers for East Africa since 2003. Most funeral homes in the U.S. and Europe are legally obliged to remove pacemakers from the dead before cremation. “Most pacemakers survive their owners,“ says Israel. He sterilizes the pacemakers and finds them a new life in East Africa. Studies show that reused pacemakers carry no greater risk for the patients than new ones.
In the U.S., University of Michigan professor Thomas Crawford heads up a similar initiative, My Heart, Your Heart. “Each year 1 to 2 million individuals worldwide die due to a lack of access to pacemakers and defibrillators,” the organization notes on its website. The nonprofit was founded in 2009, but it took four years for the doctors to get permission from the FDA to export pacemakers. Since receiving permission, the organization has sent dozens of devices to the Philippines, Haiti, Venezuela, Kenya, Sierra Leone and Ukraine. “We were the first doctors ever to implant a pacemaker in Sierra Leone in 2018,” says Crawford, who has traveled extensively to most of the recipient countries.
Even individuals can donate their pacemakers; the organization offers a prepaid envelope. “My mother recently passed and she donated her device,” says Tina Alexandris-Souphis, one of the doctors at University of Michigan who collaborates on My Heart, Your Heart. The organization works with World Medical Relief and the U.K. based charity Pace4Life to maintain a registry of the most urgent patients and send devices to where they are needed the most.
My Heart, Your Heart is also conducting a randomized controlled trial to provide further evidence that reused pacemakers pose no additional risk. “Our vision is that we establish this is safe and create a blueprint for organizations around the world to safely reuse these devices instead of them being thrown in the trash,” says Eva Kline Rogers, the program’s coordinator. “If we get this right, we have lots of devices we can implant, hips and knees, etc. Where this will lead is limitless.” She points out that in addition to receiving the donated devices, the doctors in the global South also benefit from the expertise of renowned cardiologists, such as Crawford, who sometimes advise them in complex cases.
And Adrian Baranchuk, a Canadian doctor at the Kingston General Hospital at the Queen’s University, regularly travels through South America with his “cardiology van” to help villagers in remote areas with heart problems.
Israel says that he’s been accused of racism, in thinking that these pacemakers are suitable for those in the global South - many of whom are people of color - even though officials in wealthier countries consider them to be trash. The cardiologist counters such criticism with stories about desperate need of his patients. At his first medical visit to Nairobi that he organized with a local cardiologist, six patients were waiting for him. “In Germany, they would all be considered emergencies,” Israel says. One eighty-year old grandmother had a heartrate of 18. “I’ve never before seen anything like this,” Israel exclaims. “At first I thought I couldn’t find her pulse before I realized that her heart was only beating once every three seconds.” After the surgery, she got up, dressed herself and hurriedly packed her bag, explaining she had a ton of work to accomplish. Her family was in disbelief, Israel says. “They told me she had been bedridden for five years because as soon as she tried to get up she would faint.”

Israel has been accused of racism, in thinking that these pacemakers are suitable for those in the global South even though they're considered to be trash by officials in wealthier countries. The cardiologist counters such criticism with stories about desperate need of his patients.
Carsten Israel
The hospital in Nairobi where Israel conducts the surgeries, charges patients $200 for the use of its facilities. If patients can’t afford that sum, Israel will pay it from the funds of his nonprofit. For some people, $200 far exceeds their resources. Once, a family from the extremely poor Northern region of Kenya told him they couldn’t afford the $3 for the bus ride to Nairobi. Israel suspected this was a pretense because they were afraid of the surgery and agreed to reimburse the $3, “but when they came, they were wearing rags and were so rail-thin, I understood that they really needed every cent they had for food.”
Israel is a renowned cardiologists in Germany. And yet, he considers his work in East Africa to be particularly meaningful. “Generally, most patients in Germany will get the treatment they need,” he says, “and I never before experienced that people have an illness that is easily curable but simply won’t be treated.” He also feels a heavy responsibility. Many patients have his personal cell phone and call him when they have problems or good news about how they’re doing.
Some of those progress reports come much faster than in Israel’s home country. Before he implanted a pacemaker in a tall Massai in Kenya, the man had been picked on by his family because he wouldn’t help much with the hard work on the family peanut farm. “When I examined him, he had a pulse of 40,” Israel says. “It’s a miracle he was even standing upright, let alone hauling heavy bags.” After the surgery, Israel advised his patient to stay the night for observation, but the patient couldn’t wait to leave. Two hours later, he returned, covered in sweat. He’d been running sprints with his brothers to test the new device. Israel shakes his head. In Germany, it would be unthinkable for a patient to engage in athletics immediately after surgery. But the patient was exuberant: “I was the fastest!”
The success stories are notable partly because the challenges remain so steep. In Zambia, for instance, there is a single cardiologist; she determined to become one after losing her younger sister to an easily curable heart disease. Often, the hospitals not only lack pacemakers but also sterile surgery equipment, antibiotics and other essential material. Therefore, Israel and his team import everything they need for the surgeries, including medication. If necessary, they improvise. “I’ve done surgery with a desk lamp hanging from the ceiling by threads,” Israel says. He already knows that he will need to return to Kenya in six months to replace the pacemaker of one of his patients and replace the batteries in others. If he doesn’t travel, lives are at risk.These technologies may help more animals and plants survive climate change
As the climate changes, the ripples will reach everywhere. Better data is needed for both plants and animals, and scientists are looking for genes that could allow crops to survive.
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are making us more vulnerable to infectious diseases by land and by sea - and how scientists are working on solutions.
Along the west coast of South Florida and the Keys, Florida Bay is a nursery for young Caribbean spiny lobsters, a favorite local delicacy. Growing up in small shallow basins, they are especially vulnerable to warmer, more saline water. Climate change has brought tidal floods, bleached coral reefs and toxic algal blooms to the state, and since the 1990s, the population of the Caribbean spiny lobster has dropped some 20 percent, diminishing an important food for snapper, grouper, and herons, as well as people. In 1999, marine ecologist Donald Behringer discovered the first known virus among lobsters, Panulirus argus virus—about a quarter of juveniles die from it before they mature.
“When the water is warm PaV1 progresses much more quickly,” says Behringer, who is based at the Emerging Pathogens Institute at the University of Florida in Gainesville.
Caribbean spiny lobsters are only one example of many species that are struggling in the era of climate change, both at sea and on land. As the oceans heat up, absorbing greenhouse gases and growing more acidic, marine diseases are emerging at an accelerated rate. Marine creatures are migrating to new places, and carrying pathogens with them. The latest grim report in the journal Science, states that if global warming continues at the current rate, the extinction of marine species will rival the Permian–Triassic extinction, sometimes called the “Great Dying,” when volcanoes poisoned the air and wiped out as much as 90 percent of all marine life 252 million years ago.
Similarly, on land, climate change has exposed wildlife, trees and crops to new or more virulent pathogens. Warming environments allow fungi, bacteria, viruses and infectious worms to proliferate in new species and locations or become more virulent. One paper modeling records of nearly 1,400 wildlife species projects that parasites will double by 2070 in the far north and in high-altitude places. Right now, we are seeing the effects most clearly on the fringes—along the coasts, up north and high in the mountains—but as the climate continues changing, the ripples will reach everywhere.
Few species are spared
On the Hawaiian Islands, mosquitoes are killing more songbirds. The dusky gray akikiki of Kauai and the chartreuse-yellow kiwikiu of Maui could vanish in two years, under assault from mosquitoes bearing avian malaria, according to a University of Hawaiʻi 2022 report. Previously, the birds could escape infection by roosting high in the cold mountains, where the pests couldn’t thrive, but climate change expanded the range of the mosquito and narrowed theirs.
Likewise, as more midge larvae survive over warm winters and breed better during drier summers, they bite more white-tailed deer, spreading often-fatal epizootic hemorrhagic disease. Especially in northern regions of the globe, climate change brings the threat of midges carrying blue tongue disease, a virus, to sheep and other animals. Tick-borne diseases like encephalitis and Lyme disease may become a greater threat to animals and perhaps humans.
"If you put all your eggs in one basket and then a pest comes a long, then you are more vulnerable to those risks," says Mehroad Ehsani, managing director of the food initiative in Africa for the Rockefeller Foundation. "Research is needed on resilient, climate smart, regenerative agriculture."
In the “thermal mismatch” theory of wildlife disease, cold-adapted species are at greater risk when their habitats warm, and warm-adapted species suffer when their habitats cool. Mammals can adjust their body temperature to adapt to some extent. Amphibians, fish and insects that cannot regulate body temperatures may be at greater risk. Many scientists see amphibians, especially, as canaries in the coalmine, signaling toxicity.
Early melting ice can foster disease. Climate models predict that the spring thaw will come ever-earlier in the lakes of the French Pyrenees, for instance, which traditionally stayed frozen for up to half the year. The tadpoles of the midwife toad live under the ice, where they are often infected with amphibian chytrid fungus. When a seven-year study tracked the virus in three species of amphibians in Pyrenees’s Lac Arlet, the research team found that, the earlier the spring thaw arrived, the more infection rates rose in common toads— , while remaining high among the midwife toads. But the team made another sad discovery: with early thaws, the common frog, which was thought to be free of the disease in Europe, also became infected with the fungus and died in large numbers.
Changing habitats affect animal behavior. Normally, spiny lobsters rely on chemical cues to avoid predators and sick lobsters. New conditions may be hampering their ability to “social distance”—which may help PaV1 spread, Behringer’s research suggests. Migration brings other risks. In April 2022, an international team led by scientists at Georgetown University announced the first comprehensive overview, published in the journal Nature, of how wild mammals under pressure from a changing climate may mingle with new populations and species—giving viruses a deadly opportunity to jump between hosts. Droughts, for example, will push animals to congregate at the few places where water remains.
Plants face threats also. At the timberline of the cold, windy, snowy mountains of the U.S. west, whitebark pine forests are facing a double threat, from white pine blister rust, a fungal disease, and multiplying pine beetles. “If we do nothing, we will lose the species,” says Robert Keane, a research ecologist for the U.S. Forest Service, based in Missoula, Montana. That would be a huge shift, he explains: “It’s a keystone species. There are over 110 animals that depend on it, many insects, and hundreds of plants.” In the past, beetle larvae would take two years to complete their lifecycle, and many died in frost. “With climate change, we're seeing more and more beetles survive, and sometimes the beetle can complete its lifecycle in one year,” he says.
Quintessential crops are under threat too
As some pathogens move north and new ones develop, they pose novel threats to the crops humans depend upon. This is already happening to wheat, coffee, bananas and maize.
Breeding against wheat stem rust, a fungus long linked to famine, was a key success in the mid-20th century Green Revolution, which brought higher yields around the world. In 2013, wheat stem rust reemerged in Germany after decades of absence. It ravaged both bread and durum wheat in Sicily in 2016 and has spread as far as England and Ireland. Wheat blast disease, caused by a different fungus, appeared in Bangladesh in 2016, and spread to India, the world’s second largest producer of wheat.
Insects, moths, worms, and coffee leaf rust—a fungus now found in all coffee-growing countries—threaten the livelihoods of millions of people who grow coffee, as well as everybody’s cup of joe. More heat, more intense rain, and higher humidity have allowed coffee leaf rust to cycle more rapidly. It has grown exponentially, overcoming the agricultural chemicals that once kept it under control.
To identify new diseases and fine-tune crops for resistance, scientists are increasingly relying on genomic tools.
Tar spot, a fungus native to Latin America that can cut corn production in half, has emerged in highland areas of Central Mexico and parts of the U.S.. Meanwhile, maize lethal necrosis disease has spread to multiple countries in Africa, notes Mehrdad Ehsani, Managing Director for the Food Initiative in Africa of the Rockefeller Foundation. The Cavendish banana, which most people eat today, was bred to be resistant to the fungus Panama 1. Now a new fungus, Panama 4, has emerged on every continent–including areas of Latin America that rely on the Cavendish for their income, reported a recent story in the Guardian. New threats are poised to emerge. Potato growers in the Andes Mountains have been shielded from disease because of colder weather at high altitude, but temperature fluxes and warming weather are expected to make this crop vulnerable to potato blight, found plant pathologist Erica Goss, at the Emerging Pathogens Institute.
Science seeks solutions
To protect food supplies in the era of climate change, scientists are calling for integrated global surveillance systems for crop disease outbreaks. “You can imagine that a new crop variety that is drought-tolerant could be susceptible to a pathogen that previous varieties had some resistance against,” Goss says. “Or a country suffers from a calamitous weather event, has to import seed from another country, and that seed is contaminated with a new pathogen or more virulent strain of an existing pathogen.” Researchers at the John Innes Center in Norwich and Aarhus University in Denmark have established ways to monitor wheat rust, for example.
Better data is essential, for both plants and animals. Historically, models of climate change predicted effects on plant pathogens based on mean temperatures, and scientists tracked plant responses to constant temperatures, explains Goss. “There is a need for more realistic tests of the effects of changing temperatures, particularly changes in daily high and low temperatures on pathogens,” she says.
To identify new diseases and fine-tune crops for resistance, scientists are increasingly relying on genomic tools. Goss suggests factoring the impact of climate change into those tools. Genomic efforts to select soft red winter wheat that is resistant to Fusarium head blight (FHB), a fungus that plagues farmers in the Southeastern U.S., have had early success. But temperature changes introduce a new factor.
A fundamental solution would be to bring back diversification in farming, says Ehsani. Thousands of plant species are edible, yet we rely on a handful. Wild relatives of domesticated crops are a store of possibly useful genes that may confer resistance to disease. The same is true for livestock. “If you put all your eggs in one basket and then a pest comes along, then you are more vulnerable to those risks. Research is needed on resilient, climate smart, regenerative agriculture,” Ehsani says.
Jonathan Sleeman, director of the U.S. Geological Survey National Wildlife Health Center, has called for data on wildlife health to be systematically collected and integrated with climate and other variables because more comprehensive data will result in better preventive action. “We have focused on detecting diseases,” he says, but a more holistic strategy would apply human public health concepts to assuring animal wellbeing. (For example, one study asked experts to draw a diagram of relationships of all the factors affecting the health of a particular group of caribou.) We must not take the health of plants and animals for granted, because their vulnerability inevitably affects us too, Sleeman says. “We need to improve the resilience of wildlife populations so they can withstand the impact of climate change.”