AI and you: Is the promise of personalized nutrition apps worth the hype?

Personalized nutrition apps could provide valuable data to people trying to eat healthier, though more research must be done to show effectiveness.
As a type 2 diabetic, Michael Snyder has long been interested in how blood sugar levels vary from one person to another in response to the same food, and whether a more personalized approach to nutrition could help tackle the rapidly cascading levels of diabetes and obesity in much of the western world.
Eight years ago, Snyder, who directs the Center for Genomics and Personalized Medicine at Stanford University, decided to put his theories to the test. In the 2000s continuous glucose monitoring, or CGM, had begun to revolutionize the lives of diabetics, both type 1 and type 2. Using spherical sensors which sit on the upper arm or abdomen – with tiny wires that pierce the skin – the technology allowed patients to gain real-time updates on their blood sugar levels, transmitted directly to their phone.
It gave Snyder an idea for his research at Stanford. Applying the same technology to a group of apparently healthy people, and looking for ‘spikes’ or sudden surges in blood sugar known as hyperglycemia, could provide a means of observing how their bodies reacted to an array of foods.
“We discovered that different foods spike people differently,” he says. “Some people spike to pasta, others to bread, others to bananas, and so on. It’s very personalized and our feeling was that building programs around these devices could be extremely powerful for better managing people’s glucose.”
Unbeknown to Snyder at the time, thousands of miles away, a group of Israeli scientists at the Weizmann Institute of Science were doing exactly the same experiments. In 2015, they published a landmark paper which used CGM to track the blood sugar levels of 800 people over several days, showing that the biological response to identical foods can vary wildly. Like Snyder, they theorized that giving people a greater understanding of their own glucose responses, so they spend more time in the normal range, may reduce the prevalence of type 2 diabetes.
The commercial potential of such apps is clear, but the underlying science continues to generate intriguing findings.
“At the moment 33 percent of the U.S. population is pre-diabetic, and 70 percent of those pre-diabetics will become diabetic,” says Snyder. “Those numbers are going up, so it’s pretty clear we need to do something about it.”
Fast forward to 2022,and both teams have converted their ideas into subscription-based dietary apps which use artificial intelligence to offer data-informed nutritional and lifestyle recommendations. Snyder’s spinoff, January AI, combines CGM information with heart rate, sleep, and activity data to advise on foods to avoid and the best times to exercise. DayTwo–a start-up which utilizes the findings of Weizmann Institute of Science–obtains microbiome information by sequencing stool samples, and combines this with blood glucose data to rate ‘good’ and ‘bad’ foods for a particular person.
“CGMs can be used to devise personalized diets,” says Eran Elinav, an immunology professor and microbiota researcher at the Weizmann Institute of Science in addition to serving as a scientific consultant for DayTwo. “However, this process can be cumbersome. Therefore, in our lab we created an algorithm, based on data acquired from a big cohort of people, which can accurately predict post-meal glucose responses on a personal basis.”
The commercial potential of such apps is clear. DayTwo, who market their product to corporate employers and health insurers rather than individual consumers, recently raised $37 million in funding. But the underlying science continues to generate intriguing findings.
Last year, Elinav and colleagues published a study on 225 individuals with pre-diabetes which found that they achieved better blood sugar control when they followed a personalized diet based on DayTwo’s recommendations, compared to a Mediterranean diet. The journal Cell just released a new paper from Snyder’s group which shows that different types of fibre benefit people in different ways.
“The idea is you hear different fibres are good for you,” says Snyder. “But if you look at fibres they’re all over the map—it’s like saying all animals are the same. The responses are very individual. For a lot of people [a type of fibre called] arabinoxylan clearly reduced cholesterol while the fibre inulin had no effect. But in some people, it was the complete opposite.”

Eight years ago, Stanford's Michael Snyder began studying how continuous glucose monitors could be used by patients to gain real-time updates on their blood sugar levels, transmitted directly to their phone.
The Snyder Lab, Stanford Medicine
Because of studies like these, interest in precision nutrition approaches has exploded in recent years. In January, the National Institutes of Health announced that they are spending $170 million on a five year, multi-center initiative which aims to develop algorithms based on a whole range of data sources from blood sugar to sleep, exercise, stress, microbiome and even genomic information which can help predict which diets are most suitable for a particular individual.
“There's so many different factors which influence what you put into your mouth but also what happens to different types of nutrients and how that ultimately affects your health, which means you can’t have a one-size-fits-all set of nutritional guidelines for everyone,” says Bruce Y. Lee, professor of health policy and management at the City University of New York Graduate School of Public Health.
With the falling costs of genomic sequencing, other precision nutrition clinical trials are choosing to look at whether our genomes alone can yield key information about what our diets should look like, an emerging field of research known as nutrigenomics.
The ASPIRE-DNA clinical trial at Imperial College London is aiming to see whether particular genetic variants can be used to classify individuals into two groups, those who are more glucose sensitive to fat and those who are more sensitive to carbohydrates. By following a tailored diet based on these sensitivities, the trial aims to see whether it can prevent people with pre-diabetes from developing the disease.
But while much hope is riding on these trials, even precision nutrition advocates caution that the field remains in the very earliest of stages. Lars-Oliver Klotz, professor of nutrigenomics at Friedrich-Schiller-University in Jena, Germany, says that while the overall goal is to identify means of avoiding nutrition-related diseases, genomic data alone is unlikely to be sufficient to prevent obesity and type 2 diabetes.
“Genome data is rather simple to acquire these days as sequencing techniques have dramatically advanced in recent years,” he says. “However, the predictive value of just genome sequencing is too low in the case of obesity and prediabetes.”
Others say that while genomic data can yield useful information in terms of how different people metabolize different types of fat and specific nutrients such as B vitamins, there is a need for more research before it can be utilized in an algorithm for making dietary recommendations.
“I think it’s a little early,” says Eileen Gibney, a professor at University College Dublin. “We’ve identified a limited number of gene-nutrient interactions so far, but we need more randomized control trials of people with different genetic profiles on the same diet, to see whether they respond differently, and if that can be explained by their genetic differences.”
Some start-ups have already come unstuck for promising too much, or pushing recommendations which are not based on scientifically rigorous trials. The world of precision nutrition apps was dubbed a ‘Wild West’ by some commentators after the founders of uBiome – a start-up which offered nutritional recommendations based on information obtained from sequencing stool samples –were charged with fraud last year. The weight-loss app Noom, which was valued at $3.7 billion in May 2021, has been criticized on Twitter by a number of users who claimed that its recommendations have led to them developed eating disorders.
With precision nutrition apps marketing their technology at healthy individuals, question marks have also been raised about the value which can be gained through non-diabetics monitoring their blood sugar through CGM. While some small studies have found that wearing a CGM can make overweight or obese individuals more motivated to exercise, there is still a lack of conclusive evidence showing that this translates to improved health.
However, independent researchers remain intrigued by the technology, and say that the wealth of data generated through such apps could be used to help further stratify the different types of people who become at risk of developing type 2 diabetes.
“CGM not only enables a longer sampling time for capturing glucose levels, but will also capture lifestyle factors,” says Robert Wagner, a diabetes researcher at University Hospital Düsseldorf. “It is probable that it can be used to identify many clusters of prediabetic metabolism and predict the risk of diabetes and its complications, but maybe also specific cardiometabolic risk constellations. However, we still don’t know which forms of diabetes can be prevented by such approaches and how feasible and long-lasting such self-feedback dietary modifications are.”
Snyder himself has now been wearing a CGM for eight years, and he credits the insights it provides with helping him to manage his own diabetes. “My CGM still gives me novel insights into what foods and behaviors affect my glucose levels,” he says.
He is now looking to run clinical trials with his group at Stanford to see whether following a precision nutrition approach based on CGM and microbiome data, combined with other health information, can be used to reverse signs of pre-diabetes. If it proves successful, January AI may look to incorporate microbiome data in future.
“Ultimately, what I want to do is be able take people’s poop samples, maybe a blood draw, and say, ‘Alright, based on these parameters, this is what I think is going to spike you,’ and then have a CGM to test that out,” he says. “Getting very predictive about this, so right from the get go, you can have people better manage their health and then use the glucose monitor to help follow that.”
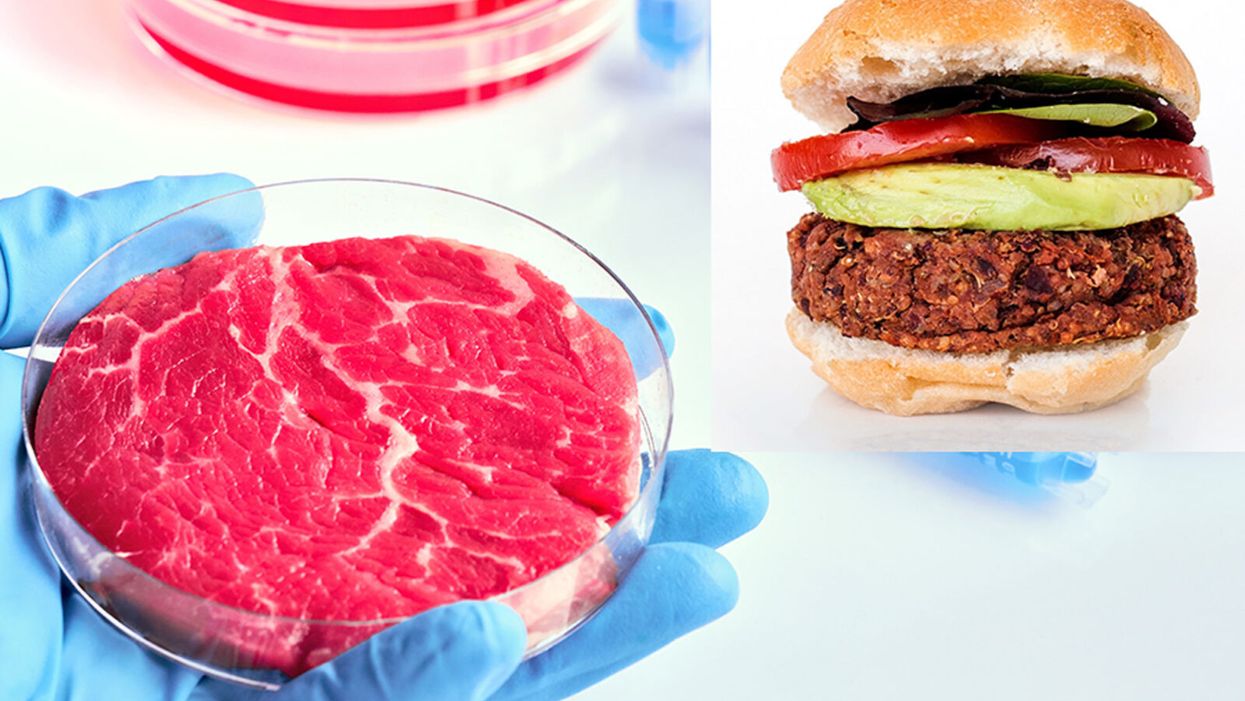
While lab-grown meat is not yet ready to crisis-proof the food supply chain, it offers key benefits that could make it an important solution beyond this pandemic.
The coronavirus pandemic exposed significant weaknesses in the country's food supply chain. Grocery store meat counters were bare. Transportation interruptions influenced supply. Finding beef, poultry, and pork at the store has been, in some places, as challenging as finding toilet paper.
In traditional agriculture models, it takes at least three months to raise chicken, six to nine months for pigs, and 18 months for cattle.
It wasn't a lack of supply -- millions of animals were in the pipeline.
"There's certainly enough food out there, but it can't get anywhere because of the way our system is set up," said Amy Rowat, an associate professor of integrative biology and physiology at UCLA. "Having a more self-contained, self-sufficient way to produce meat could make the supply chain more robust."
Cultured meat could be one way of making the meat supply chain more resilient despite disruptions due to pandemics such as COVID-19. But is the country ready to embrace lab-grown food?
According to a Good Food Institute study, GenZ is almost twice as likely to embrace meat alternatives for reasons related to social and environmental awareness, even prior to the pandemic. That's because this group wants food choices that reflect their values around food justice, equity, and animal welfare.
Largely, the interest in protein alternatives has been plant-based foods. However, factors directly related to COVID-19 may accelerate consumer interest in the scaling up of cell-grown products, according to Liz Specht, the associate director of science and technology at The Good Food Institute. The latter is a nonprofit organization that supports scientists, investors, and entrepreneurs working to develop food alternatives to conventional animal products.
While lab-grown food isn't ready yet to definitively crisis-proof the food supply chain, experts say it offers promise.
Matching Supply and Demand
Companies developing cell-grown meat claim it can take as few as two months to develop a cell into an edible product, according to Anthony Chow, CFA at Agronomics Limited, an investment company focused on meat alternatives. Tissue is taken from an animal and placed in a culture that contains nutrients and proteins the cells need to grow and expand. He cites a Good Food Institute report that claims a 2.5-millimeter sample can grow three and a half tons of meat in 40 days, allowing for exponential growth when needed.
In traditional agriculture models, it takes at least three months to raise chicken, six to nine months for pigs, and 18 months for cattle. To keep enough maturing animals in the pipeline, farms must plan the number of animals to raise months -- even years -- in advance. Lab-grown meat advocates say that because cultured meat supplies can be flexible, it theoretically allows for scaling up or down in significantly less time.
"Supply and demand has drastically changed in some way around the world and cultivated meat processing would be able to adapt much quicker than conventional farming," Chow said.
Scaling Up
Lab-grown meat may provide an eventual solution, but not in the immediate future, said Paul Mozdziak, a professor of physiology at North Carolina State University who researches animal cell culture techniques, transgenic animal production, and muscle biology.
"The challenge is in culture media," he said. "It's going to take some innovation to get the cells to grow at quantities that are going to be similar to what you can get from an animal. These are questions that everybody in the space is working on."
Chow says some of the most advanced cultured meat companies, such as BlueNal, anticipate introducing products to the market midway through next year. However, he thinks COVID-19 has slowed the process. Once introduced, they will be at a premium price, most likely available at restaurants before they hit grocery store shelves.
"I think in five years' time it will be in a different place," he said. "I don't think that this will have relevance for this pandemic, but certainly beyond that."
"Plant-based meats may be perceived as 'alternatives' to meat, whereas lab-grown meat is producing the same meat, just in a much more efficient manner, without the environmental implications."
Of course, all the technological solutions in the world won't solve the problem unless people are open-minded about embracing them. At least for now, a lab-grown burger or bluefin tuna might still be too strange for many people, especially in the U.S.
For instance, a 2019 article published by "Frontiers in Sustainable Food Systems" reflects results from a study of 3,030 consumers showing that 29 percent of U.S. customers, 59 percent of Chinese consumers, and 56 percent of Indian consumers were either 'very' or 'extremely likely' to try cultivated meat.
"Lab-grown meat is genuine meat, at the cellular level, and therefore will match conventional meat with regard to its nutritional content and overall sensory experience. It could be argued that plant-based meat will never be able to achieve this," says Laura Turner, who works with Chow at Agronomics Limited. "Plant-based meats may be perceived as 'alternatives' to meat, whereas lab-grown meat is producing the same meat, just in a much more efficient manner, without the environmental implications."
A Solution Beyond This Pandemic
The coronavirus has done more than raise awareness of the fragility of food supply chains. It has also been a wakeup call for consumers and policy makers that it is time to radically rethink our meat, Specht says. Those factors have elevated the profile of lab-grown meat.
"I think the economy is getting a little bit more steam and if I was an investor, I would be getting excited about it," adds Mozdziak.
Beyond crises, Mozdziak explains that as affluence continues to increase globally, meat consumption increases exponentially. Yet farm animals can only grow so quickly and traditional farming won't be able to keep up.
"Even Tyson is saying that by 2050, there's not going to be enough capacity in the animal meat space to meet demand," he notes. "If we don't look at some innovative technologies, how are we going to overcome that?"
A COVID-19 moonshot program challenged chemists around the world to submit ideas for how best to design a drug to target the virus.
By mid-March, Alpha Lee was growing restless. A pioneer of AI-driven drug discovery, Lee leads a team of researchers at the University of Cambridge, but his lab had been closed amidst the government-initiated lockdowns spreading inexorably across Europe.
If the Moonshot proves successful, they hope it could serve as a future benchmark for finding new medicines for chronic diseases.
Having spoken to his collaborators across the globe – many of whom were seeing their own experiments and research projects postponed indefinitely due to the pandemic – he noticed a similar sense of frustration and helplessness in the face of COVID-19.
While there was talk of finding a novel treatment for the virus, Lee was well aware the process was likely to be long and laborious. Traditional methods of drug discovery risked suffering the same fate as the efforts to find a cure for SARS in the early 2000, which took years and were ultimately abandoned long before a drug ever reached the market.
To avoid such an outcome, Lee was convinced that global collaboration was required. Together with a collection of scientists in the UK, US and Israel, he launched the 'COVID Moonshot' – a project which encouraged chemists worldwide to share their ideas for potential drug designs. If the Moonshot proves successful, they hope it could serve as a future benchmark for finding new medicines for chronic diseases.
Solving a Complex Jigsaw
In February, ShanghaiTech University published the first detailed snapshots of the SARS-CoV-2 coronavirus's proteins using a technique called X-ray crystallography. In particular, they revealed a high-resolution profile of the virus's main protease – the part of its structure that enables it to replicate inside a host – and the main drug target. The images were tantalizing.
"We could see all the tiny pieces sitting in the structure like pieces of a jigsaw," said Lee. "All we needed was for someone to come up with the best idea of joining these pieces together with a drug. Then you'd be left with a strong molecule which sits in the protease, and stops it from working, killing the virus in the process."
Normally, ideas for how best to design such a drug would be kept as carefully guarded secrets within individual labs and companies due to their potential value. But as a result, the steady process of trial and error to reach an optimum design can take years to come to fruition.
However, given the scale of the global emergency, Lee felt that the scientific community would be open to collective brainstorming on a mass scale. "Big Pharma usually wouldn't necessarily do this, but time is of the essence here," he said. "It was a case of, 'Let's just rethink every drug discovery stage to see -- ok, how can we go as fast as we can?'"
On March 13, he launched the COVID moonshot, calling for chemists around the globe to come up with the most creative ideas they could think of, on their laptops at home. No design was too weird or wacky to be considered, and crucially nothing would be patented. The entire project would be done on a not-for-profit basis, meaning that any drug that makes it to market will have been created simply for the good of humanity.
It caught fire: Within just two weeks, more than 2,300 potential drug designs had been submitted. By the middle of July, over 10,000 had been received from scientists around the globe.
The Road Toward Clinical Trials
With so many designs to choose from, the team has been attempting to whittle them down to a shortlist of the most promising. Computational drug discovery experts at Diamond and the Weizmann Institute of Science in Rehovot, Israel, have enabled the Moonshot team to develop algorithms for predicting how quick and easy each design would be to make, and to predict how well each proposed drug might bind to the virus in real life.
The latter is an approach known as computational covalent docking and has previously been used in cancer research. "This was becoming more popular even before COVID-19, with several covalent drugs approved by the FDA in recent years," said Nir London, professor of organic chemistry at the Weizmann Institute, and one of the Moonshot team members. "However, all of these were for oncology. A covalent drug against SARS-CoV-2 will certainly highlight covalent drug-discovery as a viable option."
Through this approach, the team have selected 850 compounds to date, which they have manufactured and tested in various preclinical trials already. Fifty of these compounds - which appear to be especially promising when it comes to killing the virus in a test tube – are now being optimized further.
Lee is hoping that at least one of these potential drugs will be shown to be effective in curing animals of COVID-19 within the next six months, a step that would allow the Moonshot team to reach out to potential pharmaceutical partners to test their compounds in humans.
Future Implications
If the project does succeed, some believe it could open the door to scientific crowdsourcing as a future means of generating novel medicine ideas for other diseases. Frank von Delft, professor of protein science and structural biology at the University of Oxford's Nuffield Department of Medicine, described it as a new form of 'citizen science.'
"There's a vast resource of expertise and imagination that is simply dying to be tapped into," he said.
Others are slightly more skeptical, pointing out that the uniqueness of the current crisis has meant that many scientists were willing to contribute ideas without expecting any future compensation in return. This meant that it was easy to circumvent the traditional hurdles that prevent large-scale global collaborations from happening – namely how to decide who will profit from the final product and who will hold the intellectual property (IP) rights.
"I think it is too early to judge if this is a viable model for future drug discovery," says London. "I am not sure that without the existential threat we would have seen so many contributions, and so many people and institutions willing to waive compensation and future royalties. Many scientists found themselves at home, frustrated that they don't have a way to contribute to the fight against COVID-19, and this project gave them an opportunity. Plus many can get behind the fact that this project has no associated IP and no one will get rich off of this effort. This breaks down a lot of the typical barriers and red-tape for wider collaboration."
"If a drug would sprout from one of these crowdsourced ideas, it would serve as a very powerful argument to consider this mode of drug discovery further in the future."
However the Moonshot team believes that if they can succeed, it will at the very least send a strong statement to policy makers and the scientific community that greater efforts should be made to make such large-scale collaborations more feasible.
"All across the scientific world, we've seen unprecedented adoption of open-science, collaboration and collegiality during this crisis, perhaps recognizing that only a coordinated global effort could address this global challenge," says London. "If a drug would sprout from one of these crowdsourced ideas, it would serve as a very powerful argument to consider this mode of drug discovery further in the future."
[An earlier version of this article was published on June 8th, 2020 as part of a standalone magazine called GOOD10: The Pandemic Issue. Produced as a partnership among LeapsMag, The Aspen Institute, and GOOD, the magazine is available for free online.]