AI and you: Is the promise of personalized nutrition apps worth the hype?

Personalized nutrition apps could provide valuable data to people trying to eat healthier, though more research must be done to show effectiveness.
As a type 2 diabetic, Michael Snyder has long been interested in how blood sugar levels vary from one person to another in response to the same food, and whether a more personalized approach to nutrition could help tackle the rapidly cascading levels of diabetes and obesity in much of the western world.
Eight years ago, Snyder, who directs the Center for Genomics and Personalized Medicine at Stanford University, decided to put his theories to the test. In the 2000s continuous glucose monitoring, or CGM, had begun to revolutionize the lives of diabetics, both type 1 and type 2. Using spherical sensors which sit on the upper arm or abdomen – with tiny wires that pierce the skin – the technology allowed patients to gain real-time updates on their blood sugar levels, transmitted directly to their phone.
It gave Snyder an idea for his research at Stanford. Applying the same technology to a group of apparently healthy people, and looking for ‘spikes’ or sudden surges in blood sugar known as hyperglycemia, could provide a means of observing how their bodies reacted to an array of foods.
“We discovered that different foods spike people differently,” he says. “Some people spike to pasta, others to bread, others to bananas, and so on. It’s very personalized and our feeling was that building programs around these devices could be extremely powerful for better managing people’s glucose.”
Unbeknown to Snyder at the time, thousands of miles away, a group of Israeli scientists at the Weizmann Institute of Science were doing exactly the same experiments. In 2015, they published a landmark paper which used CGM to track the blood sugar levels of 800 people over several days, showing that the biological response to identical foods can vary wildly. Like Snyder, they theorized that giving people a greater understanding of their own glucose responses, so they spend more time in the normal range, may reduce the prevalence of type 2 diabetes.
The commercial potential of such apps is clear, but the underlying science continues to generate intriguing findings.
“At the moment 33 percent of the U.S. population is pre-diabetic, and 70 percent of those pre-diabetics will become diabetic,” says Snyder. “Those numbers are going up, so it’s pretty clear we need to do something about it.”
Fast forward to 2022,and both teams have converted their ideas into subscription-based dietary apps which use artificial intelligence to offer data-informed nutritional and lifestyle recommendations. Snyder’s spinoff, January AI, combines CGM information with heart rate, sleep, and activity data to advise on foods to avoid and the best times to exercise. DayTwo–a start-up which utilizes the findings of Weizmann Institute of Science–obtains microbiome information by sequencing stool samples, and combines this with blood glucose data to rate ‘good’ and ‘bad’ foods for a particular person.
“CGMs can be used to devise personalized diets,” says Eran Elinav, an immunology professor and microbiota researcher at the Weizmann Institute of Science in addition to serving as a scientific consultant for DayTwo. “However, this process can be cumbersome. Therefore, in our lab we created an algorithm, based on data acquired from a big cohort of people, which can accurately predict post-meal glucose responses on a personal basis.”
The commercial potential of such apps is clear. DayTwo, who market their product to corporate employers and health insurers rather than individual consumers, recently raised $37 million in funding. But the underlying science continues to generate intriguing findings.
Last year, Elinav and colleagues published a study on 225 individuals with pre-diabetes which found that they achieved better blood sugar control when they followed a personalized diet based on DayTwo’s recommendations, compared to a Mediterranean diet. The journal Cell just released a new paper from Snyder’s group which shows that different types of fibre benefit people in different ways.
“The idea is you hear different fibres are good for you,” says Snyder. “But if you look at fibres they’re all over the map—it’s like saying all animals are the same. The responses are very individual. For a lot of people [a type of fibre called] arabinoxylan clearly reduced cholesterol while the fibre inulin had no effect. But in some people, it was the complete opposite.”

Eight years ago, Stanford's Michael Snyder began studying how continuous glucose monitors could be used by patients to gain real-time updates on their blood sugar levels, transmitted directly to their phone.
The Snyder Lab, Stanford Medicine
Because of studies like these, interest in precision nutrition approaches has exploded in recent years. In January, the National Institutes of Health announced that they are spending $170 million on a five year, multi-center initiative which aims to develop algorithms based on a whole range of data sources from blood sugar to sleep, exercise, stress, microbiome and even genomic information which can help predict which diets are most suitable for a particular individual.
“There's so many different factors which influence what you put into your mouth but also what happens to different types of nutrients and how that ultimately affects your health, which means you can’t have a one-size-fits-all set of nutritional guidelines for everyone,” says Bruce Y. Lee, professor of health policy and management at the City University of New York Graduate School of Public Health.
With the falling costs of genomic sequencing, other precision nutrition clinical trials are choosing to look at whether our genomes alone can yield key information about what our diets should look like, an emerging field of research known as nutrigenomics.
The ASPIRE-DNA clinical trial at Imperial College London is aiming to see whether particular genetic variants can be used to classify individuals into two groups, those who are more glucose sensitive to fat and those who are more sensitive to carbohydrates. By following a tailored diet based on these sensitivities, the trial aims to see whether it can prevent people with pre-diabetes from developing the disease.
But while much hope is riding on these trials, even precision nutrition advocates caution that the field remains in the very earliest of stages. Lars-Oliver Klotz, professor of nutrigenomics at Friedrich-Schiller-University in Jena, Germany, says that while the overall goal is to identify means of avoiding nutrition-related diseases, genomic data alone is unlikely to be sufficient to prevent obesity and type 2 diabetes.
“Genome data is rather simple to acquire these days as sequencing techniques have dramatically advanced in recent years,” he says. “However, the predictive value of just genome sequencing is too low in the case of obesity and prediabetes.”
Others say that while genomic data can yield useful information in terms of how different people metabolize different types of fat and specific nutrients such as B vitamins, there is a need for more research before it can be utilized in an algorithm for making dietary recommendations.
“I think it’s a little early,” says Eileen Gibney, a professor at University College Dublin. “We’ve identified a limited number of gene-nutrient interactions so far, but we need more randomized control trials of people with different genetic profiles on the same diet, to see whether they respond differently, and if that can be explained by their genetic differences.”
Some start-ups have already come unstuck for promising too much, or pushing recommendations which are not based on scientifically rigorous trials. The world of precision nutrition apps was dubbed a ‘Wild West’ by some commentators after the founders of uBiome – a start-up which offered nutritional recommendations based on information obtained from sequencing stool samples –were charged with fraud last year. The weight-loss app Noom, which was valued at $3.7 billion in May 2021, has been criticized on Twitter by a number of users who claimed that its recommendations have led to them developed eating disorders.
With precision nutrition apps marketing their technology at healthy individuals, question marks have also been raised about the value which can be gained through non-diabetics monitoring their blood sugar through CGM. While some small studies have found that wearing a CGM can make overweight or obese individuals more motivated to exercise, there is still a lack of conclusive evidence showing that this translates to improved health.
However, independent researchers remain intrigued by the technology, and say that the wealth of data generated through such apps could be used to help further stratify the different types of people who become at risk of developing type 2 diabetes.
“CGM not only enables a longer sampling time for capturing glucose levels, but will also capture lifestyle factors,” says Robert Wagner, a diabetes researcher at University Hospital Düsseldorf. “It is probable that it can be used to identify many clusters of prediabetic metabolism and predict the risk of diabetes and its complications, but maybe also specific cardiometabolic risk constellations. However, we still don’t know which forms of diabetes can be prevented by such approaches and how feasible and long-lasting such self-feedback dietary modifications are.”
Snyder himself has now been wearing a CGM for eight years, and he credits the insights it provides with helping him to manage his own diabetes. “My CGM still gives me novel insights into what foods and behaviors affect my glucose levels,” he says.
He is now looking to run clinical trials with his group at Stanford to see whether following a precision nutrition approach based on CGM and microbiome data, combined with other health information, can be used to reverse signs of pre-diabetes. If it proves successful, January AI may look to incorporate microbiome data in future.
“Ultimately, what I want to do is be able take people’s poop samples, maybe a blood draw, and say, ‘Alright, based on these parameters, this is what I think is going to spike you,’ and then have a CGM to test that out,” he says. “Getting very predictive about this, so right from the get go, you can have people better manage their health and then use the glucose monitor to help follow that.”
Three Big Biotech Ideas to Watch in 2020—And Beyond
Body-on-a-chip, prime editing, and gut microbes all are poised to make a big impact in 2020.
1. Happening Now: Body-on-a-Chip Technology Is Enabling Safer Drug Trials and Better Cancer Research
Researchers have increasingly used the technology known as "lab-on-a-chip" or "organ-on-a-chip" to test the effects of pharmaceuticals, toxins, and chemicals on humans. Rather than testing on animals, which raises ethical concerns and can sometimes be inaccurate, and human-based clinical trials, which can be expensive and difficult to iterate, scientists turn to tiny, micro-engineered chips—about the size of a thumb drive.
It's possible that doctors could one day take individual cell samples and create personalized treatments, testing out any medications on the chip.
The chips are lined with living samples of human cells, which mimic the physiology and mechanical forces experienced by cells inside the human body, down to blood flow and breathing motions; the functions of organs ranging from kidneys and lungs to skin, eyes, and the blood-brain barrier.
A more recent—and potentially even more useful—development takes organ-on-a-chip technology to the next level by integrating several chips into a "body-on-a-chip." Since human organs don't work in isolation, seeing how they all react—and interact—once a foreign element has been introduced can be crucial to understanding how a certain treatment will or won't perform. Dr. Shyni Varghese, a MEDx investigator at the Duke University School of Medicine, is one of the researchers working with these systems in order to gain a more nuanced understanding of how multiple different organs react to the same stimuli.
Her lab is working on "tumor-on-a-chip" models, which can not only show the progression and treatment of cancer, but also model how other organs would react to immunotherapy and other drugs. "The effect of drugs on different organs can be tested to identify potential side effects," Varghese says. In addition, these models can help the researchers figure out how cancers grow and spread, as well as how to effectively encourage immune cells to move in and attack a tumor.
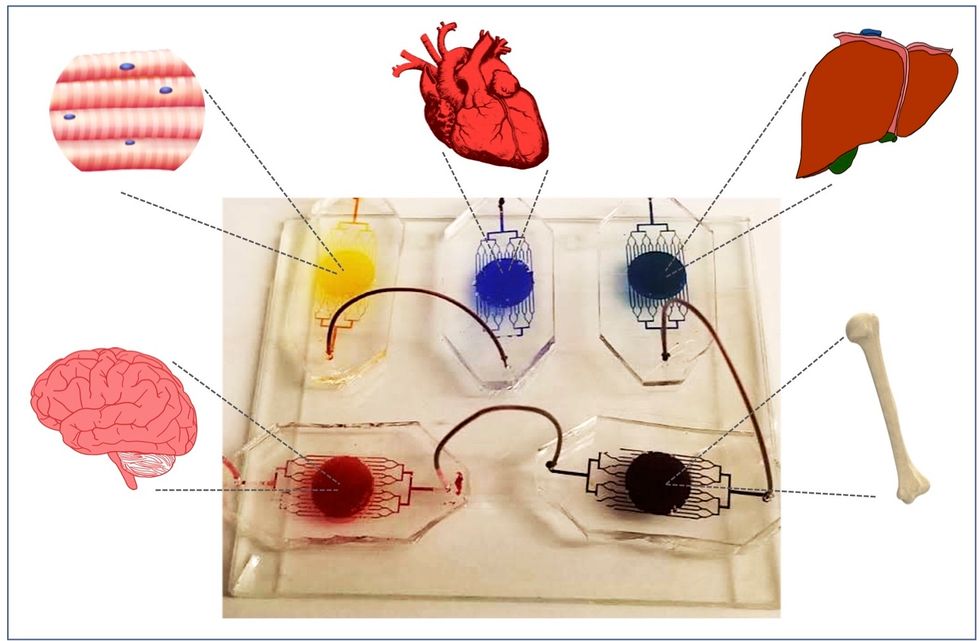
One body-on-a-chip used by Dr. Varghese's lab tracks the interactions of five organs—brain, heart, liver, muscle, and bone.
As their research progresses, Varghese and her team are looking for ways to maintain the long-term function of the engineered organs. In addition, she notes that this kind of research is not just useful for generalized testing; "organ-on-chip technologies allow patient-specific analyses, which can be used towards a fundamental understanding of disease progression," Varghese says. It's possible that doctors could one day take individual cell samples and create personalized treatments, testing out any medications on the chip for safety, efficacy, and potential side effects before writing a prescription.
2. Happening Soon: Prime Editing Will Have the Power to "Find and Replace" Disease-Causing Genes
Biochemist David Liu made industry-wide news last fall when he and his lab at MIT's Broad Institute, led by Andrew Anzalone, published a paper on prime editing: a new, more focused technology for editing genes. Prime editing is a descendant of the CRISPR-Cas9 system that researchers have been working with for years, and a cousin to Liu's previous innovation—base editing, which can make a limited number of changes to a single DNA letter at a time.
By contrast, prime editing has the potential to make much larger insertions and deletions; it also doesn't require the tweaked cells to divide in order to write the changes into the DNA, which could make it especially suitable for central nervous system diseases, like Parkinson's.
Crucially, the prime editing technique has a much higher efficiency rate than the older CRISPR system, and a much lower incidence of accidental insertions or deletions, which can make dangerous changes for a patient.
It also has a very broad potential range: according to Liu, 89% of the pathogenic mutations that have been collected in ClinVar (a public archive of human variations) could, in principle, be treated with prime editing—although he is careful to note that correcting a single genetic mutation may not be sufficient to fully treat a genetic disease.
Figuring out just how prime editing can be used most effectively and safely will be a long process, but it's already underway. The same day that Liu and his team posted their paper, they also made the basic prime editing constructs available for researchers around the world through Addgene, a plasmid repository, so that others in the scientific community can test out the technique for themselves. It might be years before human patients will see the results, and in the meantime, significant bioethical questions remain about the limits and sociological effects of such a powerful gene-editing tool. But in the long fight against genetic diseases, it's a huge step forward.
3. Happening When We Fund It: Focusing on Microbiome Health Could Help Us Tackle Social Inequality—And Vice Versa
The past decade has seen a growing awareness of the major role that the microbiome, the microbes present in our digestive tract, play in human health. Having a less-healthy microbiome is correlated with health risks like diabetes and depression, and interventions that target gut health, ranging from kombucha to fecal transplants, have cropped up with increasing frequency.
New research from the University of Maine's Dr. Suzanne Ishaq takes an even broader view, arguing that low-income and disadvantaged populations are less likely to have healthy, diverse gut bacteria, and that increasing access to beneficial microorganisms is an important juncture of social justice and public health.
"Basically, allowing people to lead healthy lives allows them to access and recruit microbes."
"Typically, having a more diverse bacterial community is associated with health, and having fewer different species is associated with illness and may leave you open to infection from bacteria that are good at exploiting opportunities," Ishaq says.
Having a healthy biome doesn't mean meeting one fixed ratio of gut bacteria, since different combinations of microbes can generate roughly similar results when they work in concert. Generally, "good" microbes are the ones that break down fiber and create the byproducts that we use for energy, or ones like lactic acid bacteria that work to make microbials and keep other bacteria in check. The microbial universe in your gut is chaotic, Ishaq says. "Microbes in your gut interact with each other, with you, with your food, or maybe they don't interact at all and pass right through you." Overall, it's tricky to name specific microbial communities that will make or break someone's health.
There are important corollaries between environment and biome health, though, which Ishaq points out: Living in urban environments reduces microbial exposure, and losing the microorganisms that humans typically source from soil and plants can reduce our adaptive immunity and ability to fight off conditions like allergies and asthma. Access to green space within cities can counteract those effects, but in the U.S. that access varies along income, education, and racial lines. Likewise, lower-income communities are more likely to live in food deserts or areas where the cheapest, most convenient food options are monotonous and low in fiber, further reducing microbial diversity.
Ishaq also suggests other areas that would benefit from further study, like the correlation between paid family leave, breastfeeding, and gut microbiota. There are technical and ethical challenges to direct experimentation with human populations—but that's not what Ishaq sees as the main impediment to future research.
"The biggest roadblock is money, and the solution is also money," she says. "Basically, allowing people to lead healthy lives allows them to access and recruit microbes."
That means investment in things we already understand to improve public health, like better education and healthcare, green space, and nutritious food. It also means funding ambitious, interdisciplinary research that will investigate the connections between urban infrastructure, housing policy, social equity, and the millions of microbes keeping us company day in and day out.
Scientists Just Started Testing a New Class of Drugs to Slow--and Even Reverse--Aging
Eliminating "zombie-like" cells, called senescent cells, may hold the key to slowing aging and its chronic diseases.
Imagine reversing the processes of aging. It's an age-old quest, and now a study from the Mayo Clinic may be the first ray of light in the dawn of that new era.
The immune system can handle a certain amount of senescence, but that capacity declines with age.
The small preliminary report, just nine patients, primarily looked at the safety and tolerability of the compounds used. But it also showed that a new class of small molecules called senolytics, which has proven to reverse markers of aging in animal studies, can work in humans.
Aging is a relentless assault of chronic diseases including Alzheimer's, cardiovascular disease, diabetes, and frailty. Developing one chronic condition strongly predicts the rapid onset of another. They pile on top of each other and impede the body's ability to respond to the next challenge.
"Potentially, by targeting fundamental aging processes, it may be possible to delay or prevent or alleviate multiple age-related conditions and many diseases as a group, instead of one at a time," says James Kirkland, the Mayo Clinic physician who led the study and is a top researcher in the growing field of geroscience, the biology of aging.
Getting Rid of "Zombie" Cells
One element common to many of the diseases is senescence, a kind of limbo or zombie-like state where cells no longer divide or perform many regular functions, but they don't die. Senescence is thought to be beneficial in that it inhibits the cancerous proliferation of cells. But in aging, the senescent cells still produce molecules that create inflammation both locally and throughout the body. It is a cycle that feeds upon itself, slowly ratcheting down normal body function and health.
Disease and harmful stimuli like radiation to treat cancer can also generate senescence, which is why young cancer patients seem to experience earlier and more rapid aging. The immune system can handle a certain amount of senescence, but that capacity declines with age. There also appears to be a threshold effect, a tipping point where senescence becomes a dominant factor in aging.
Kirkland's team used an artificial intelligence approach called machine learning to look for cell signaling networks that keep senescent cells from dying. To date, researchers have identified at least eight such signaling networks, some of which seem to be unique to a particular type of cell or tissue, but others are shared or overlap.
Then a computer search identified molecules known to disrupt these signaling pathways "and allow cells that are fully senescent to kill themselves," he explains. The process is a bit like looking for the right weapons in a video game to wipe out lingering zombie cells. But instead of swords, guns, and grenades, the list of biological tools so far includes experimental molecules, approved drugs, and natural supplements.
Treatment
"We found early on that targeting single components of those networks will only kill a very small minority of senescent cells or senescent cell types," says Kirkland. "So instead of going after one drug-one target-one disease, we're going after networks with combinations of drugs or drugs that have multiple targets. And we're going after every age-related disease."
The FDA is grappling with guidance for researchers wanting to conduct clinical trials on something as broad as aging rather than a single disease.
The large number of potential senolytic (i.e. zombie-neutralizing) compounds they identified allowed Kirkland to be choosy, "purposefully selecting drugs where the side effects profile was good...and with short elimination half-lives." The hit and run approach meant they didn't have to worry about maintaining a steady state of drugs in the body for an extended period of time. Some of the compounds they selected need only a half hour exposure to trigger the dying process in senescent cells, which can then take several days.
Work in mice has already shown impressive results in reversing diabetes, weight gain, Alzheimer's, cardiovascular disease and other conditions using senolytic agents.
That led to Kirkland's pilot study in humans with diabetes-related kidney disease using a three-day regimen of dasatinib, a kinase inhibitor first approved in 2006 to treat some forms of blood cancer, and quercetin, a flavonoid found in many plants and sold as a food supplement.
The combination was safe and well tolerated; it reduced the number of senescent cells in the belly fat of patients and restored their normal function, according to results published in September in the journal EBioMedicine. This preliminary paper was based on 9 patients in an ongoing study of 30 patients.
Kirkland cautions that these are initial and incomplete findings looking primarily at safety issues, not effectiveness. There is still much to be learned about the use of senolytics, starting with proof that they actually provide clinical benefit, and against what chronic conditions. The drug combinations, doses, duration, and frequency, not to mention potential risks all must be worked out. Additional studies of other diseases are being developed.
What's Next
Ron Kohanski, a senior administrator at the NIH National Institute on Aging (NIA), says the field of senolytics is so new that there isn't even a consensus on how to identify a senescent cell, and the FDA is grappling with guidance for researchers wanting to conduct clinical trials on something as broad as aging rather than a single disease.
Intellectual property concerns may temper the pharmaceutical industry's interest in developing senolytics to treat chronic diseases of aging. It looks like many mix-and-match combinations are possible, and many of the potential molecules identified so far are found in nature or are drugs whose patents have or will soon expire. So the ability to set high prices for such future drugs, and hence the willingness to spend money on expensive clinical trials, may be limited.
Still, Kohanski believes the field can move forward quickly because it often will include products that are already widely used and have a known safety profile. And approaches like Kirkland's hit and run strategy will minimize potential exposure and risk.
He says the NIA is going to support a number of clinical trials using these new approaches. Pharmaceutical companies may feel that they can develop a unique part of a senolytic combination regimen that will justify their investment. And if they don't, countries with socialized medicine may take the lead in supporting such research with the goal of reducing the costs of treating aging patients.

