Scientists forecast new disease outbreaks

A mosquito under the microscope.
Two years, six million deaths and still counting, scientists are searching for answers to prevent another COVID-19-like tragedy from ever occurring again. And it’s a gargantuan task.
Our disturbed ecosystems are creating more favorable conditions for the spread of infectious disease. Global warming, deforestation, rising sea levels and flooding have contributed to a rise in mosquito-borne infections and longer tick seasons. Disease-carrying animals are in closer range to other species and humans as they migrate to escape the heat. Bats are thought to have carried the SARS-CoV-2 virus to Wuhan, either directly or through another host animal, but thousands of novel viruses are lurking within other wild creatures.
Understanding how climate change contributes to the spread of disease is critical in predicting and thwarting future calamities. But the problem is that predictive models aren’t yet where they need to be for forecasting with certainty beyond the next year, as we could for weather, for instance.
The association between climate and infectious disease is poorly understood, says Irina Tezaur, a computational scientist at Sandia National Laboratories. “Correlations have been observed but it’s not known if these correlations translate to causal relationships.”
To make accurate longer-term predictions, scientists need more empirical data, multiple datasets specific to locations and diseases, and the ability to calculate risks that depend on unpredictable nature and human behavior. Another obstacle is that climate scientists and epidemiologists are not collaborating effectively, so some researchers are calling for a multidisciplinary approach, a new field called Outbreak Science.
Climate scientists are far ahead of epidemiologists in gathering essential data.
Earth System Models—combining the interactions of atmosphere, ocean, land, ice and biosphere—have been in place for two decades to monitor the effects of global climate change. These models must be combined with epidemiological and human model research, areas that are easily skewed by unpredictable elements, from extreme weather events to public environmental policy shifts.
“There is never just one driver in tracking the impact of climate on infectious disease,” says Joacim Rocklöv, a professor at the Heidelberg Institute of Global Health & Heidelberg Interdisciplinary Centre for Scientific Computing in Germany. Rocklöv has studied how climate affects vector-borne diseases—those transmitted to humans by mosquitoes, ticks or fleas. “You need to disentangle the variables to find out how much difference climate makes to the outcome and how much is other factors.” Determinants from deforestation to population density to lack of healthcare access influence the spread of disease.
Even though climate change is not the primary driver of infectious disease today, it poses a major threat to public health in the future, says Rocklöv.
The promise of predictive modeling
“Models are simplifications of a system we’re trying to understand,” says Jeremy Hess, who directs the Center for Health and the Global Environment at University of Washington in Seattle. “They’re tools for learning that improve over time with new observations.”
Accurate predictions depend on high-quality, long-term observational data but models must start with assumptions. “It’s not possible to apply an evidence-based approach for the next 40 years,” says Rocklöv. “Using models to experiment and learn is the only way to figure out what climate means for infectious disease. We collect data and analyze what already happened. What we do today will not make a difference for several decades.”
To improve accuracy, scientists develop and draw on thousands of models to cover as many scenarios as possible. One model may capture the dynamics of disease transmission while another focuses on immunity data or ocean influences or seasonal components of a virus. Further, each model needs to be disease-specific and often location-specific to be useful.
“All models have biases so it’s important to use a suite of models,” Tezaur stresses.
The modeling scientist chooses the drivers of change and parameters based on the question explored. The drivers could be increased precipitation, poverty or mosquito prevalence, for instance. Later, the scientist may need to isolate the effect of one driver so that will require another model.
There have been some related successes, such as the latest models for mosquito-borne diseases like Dengue, Zika and malaria as well as those for flu and tick-borne diseases, says Hess.
Rocklöv was part of a research team that used test data from 2018 and 2019 to identify regions at risk for West Nile virus outbreaks. Using AI, scientists were able to forecast outbreaks of the virus for the entire transmission season in Europe. “In the end, we want data-driven models; that’s what AI can accomplish,” says Rocklöv. Other researchers are making an important headway in creating a framework to predict novel host–parasite interactions.
Modeling studies can run months, years or decades. “The scientist is working with layers of data. The challenge is how to transform and couple different models together on a planetary scale,” says Jeanne Fair, a scientist at Los Alamos National Laboratory, Biosecurity and Public Health, in New Mexico.
Disease forecasting will require a significant investment into the infrastructure needed to collect data about the environment, vectors, and hosts a tall spatial and temporal resolutions.
And it’s a constantly changing picture. A modeling study in an April 2022 issue of Nature predicted that thousands of animals will migrate to cooler locales as temperatures rise. This means that various species will come into closer contact with people and other mammals for the first time. This is likely to increase the risk of emerging infectious disease transmitted from animals to humans, especially in Africa and Asia.
Other things can happen too. Global warming could precipitate viral mutations or new infectious diseases that don’t respond to antimicrobial treatments. Insecticide-resistant mosquitoes could evolve. Weather-related food insecurity could increase malnutrition and weaken people’s immune systems. And the impact of an epidemic will be worse if it co-occurs during a heatwave, flood, or drought, says Hess.
The devil is in the climate variables
Solid predictions about the future of climate and disease are not possible with so many uncertainties. Difficult-to-measure drivers must be added to the empirical model mix, such as land and water use, ecosystem changes or the public’s willingness to accept a vaccine or practice social distancing. Nor is there any precedent for calculating the effect of climate changes that are accelerating at a faster speed than ever before.
The most critical climate variables thought to influence disease spread are temperature, precipitation, humidity, sunshine and wind, according to Tezaur’s research. And then there are variables within variables. Influenza scientists, for example, found that warm winters were predictors of the most severe flu seasons in the following year.
The human factor may be the most challenging determinant. To what degree will people curtail greenhouse gas emissions, if at all? The swift development of effective COVID-19 vaccines was a game-changer, but will scientists be able to repeat it during the next pandemic? Plus, no model could predict the amount of internet-fueled COVID-19 misinformation, Fair noted. To tackle this issue, infectious disease teams are looking to include more sociologists and political scientists in their modeling.
Addressing the gaps
Currently, researchers are focusing on the near future, predicting for next year, says Fair. “When it comes to long-term, that’s where we have the most work to do.” While scientists cannot foresee how political influences and misinformation spread will affect models, they are positioned to make headway in collecting and assessing new data streams that have never been merged.
Disease forecasting will require a significant investment into the infrastructure needed to collect data about the environment, vectors, and hosts at all spatial and temporal resolutions, Fair and her co-authors stated in their recent study. For example real-time data on mosquito prevalence and diversity in various settings and times is limited or non-existent. Fair also would like to see standards set in mosquito data collection in every country. “Standardizing across the US would be a huge accomplishment,” she says.

Understanding how climate change contributes to the spread of disease is critical for thwarting future calamities.
Jeanne Fair
Hess points to a dearth of data in local and regional datasets about how extreme weather events play out in different geographic locations. His research indicates that Africa and the Middle East experienced substantial climate shifts, for example, but are unrepresented in the evidentiary database, which limits conclusions. “A model for dengue may be good in Singapore but not necessarily in Port-au-Prince,” Hess explains. And, he adds, scientists need a way of evaluating models for how effective they are.
The hope, Rocklöv says, is that in the future we will have data-driven models rather than theoretical ones. In turn, sharper statistical analyses can inform resource allocation and intervention strategies to prevent outbreaks.
Most of all, experts emphasize that epidemiologists and climate scientists must stop working in silos. If scientists can successfully merge epidemiological data with climatic, biological, environmental, ecological and demographic data, they will make better predictions about complex disease patterns. Modeling “cross talk” and among disciplines and, in some cases, refusal to release data between countries is hindering discovery and advances.
It’s time for bold transdisciplinary action, says Hess. He points to initiatives that need funding in disease surveillance and control; developing and testing interventions; community education and social mobilization; decision-support analytics to predict when and where infections will emerge; advanced methodologies to improve modeling; training scientists in data management and integrated surveillance.
Establishing a new field of Outbreak Science to coordinate collaboration would accelerate progress. Investment in decision-support modeling tools for public health teams, policy makers, and other long-term planning stakeholders is imperative, too. We need to invest in programs that encourage people from climate modeling and epidemiology to work together in a cohesive fashion, says Tezaur. Joining forces is the only way to solve the formidable challenges ahead.
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
DNA- and RNA-based electronic implants may revolutionize healthcare
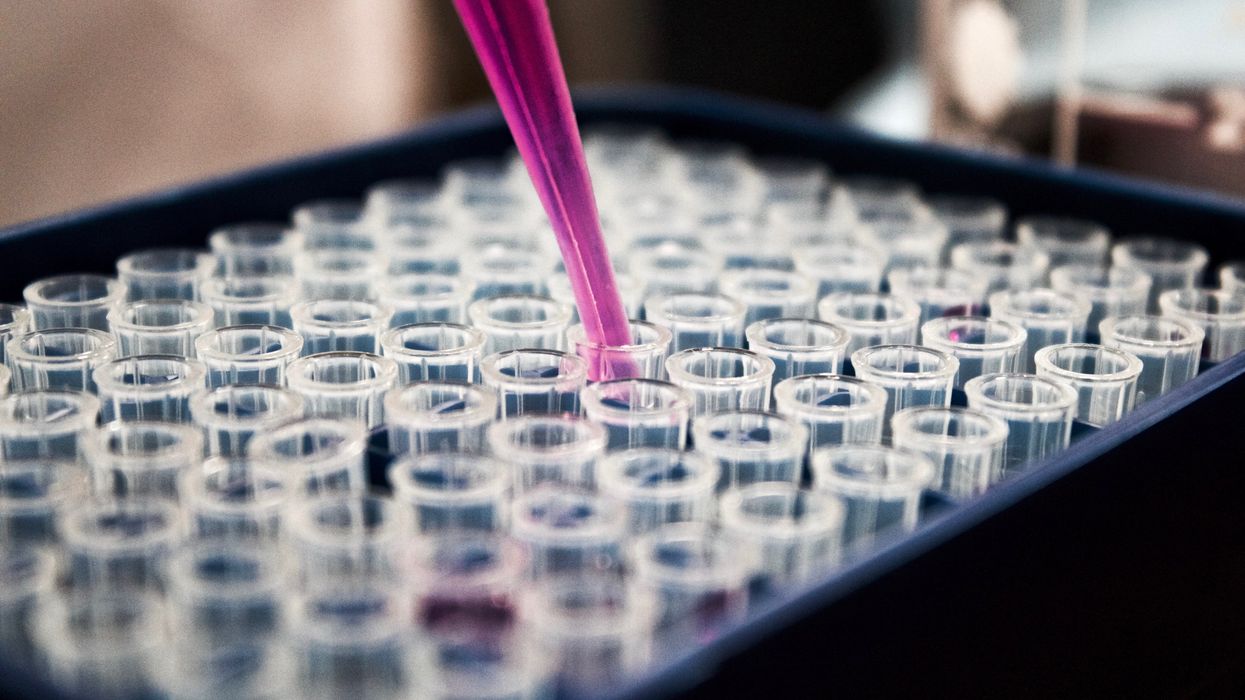
The test tubes contain tiny DNA/enzyme-based circuits, which comprise TRUMPET, a new type of electronic device, smaller than a cell.
Implantable electronic devices can significantly improve patients’ quality of life. A pacemaker can encourage the heart to beat more regularly. A neural implant, usually placed at the back of the skull, can help brain function and encourage higher neural activity. Current research on neural implants finds them helpful to patients with Parkinson’s disease, vision loss, hearing loss, and other nerve damage problems. Several of these implants, such as Elon Musk’s Neuralink, have already been approved by the FDA for human use.
Yet, pacemakers, neural implants, and other such electronic devices are not without problems. They require constant electricity, limited through batteries that need replacements. They also cause scarring. “The problem with doing this with electronics is that scar tissue forms,” explains Kate Adamala, an assistant professor of cell biology at the University of Minnesota Twin Cities. “Anytime you have something hard interacting with something soft [like muscle, skin, or tissue], the soft thing will scar. That's why there are no long-term neural implants right now.” To overcome these challenges, scientists are turning to biocomputing processes that use organic materials like DNA and RNA. Other promised benefits include “diagnostics and possibly therapeutic action, operating as nanorobots in living organisms,” writes Evgeny Katz, a professor of bioelectronics at Clarkson University, in his book DNA- And RNA-Based Computing Systems.
While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output.
Adamala’s research focuses on developing such biocomputing systems using DNA, RNA, proteins, and lipids. Using these molecules in the biocomputing systems allows the latter to be biocompatible with the human body, resulting in a natural healing process. In a recent Nature Communications study, Adamala and her team created a new biocomputing platform called TRUMPET (Transcriptional RNA Universal Multi-Purpose GatE PlaTform) which acts like a DNA-powered computer chip. “These biological systems can heal if you design them correctly,” adds Adamala. “So you can imagine a computer that will eventually heal itself.”
The basics of biocomputing
Biocomputing and regular computing have many similarities. Like regular computing, biocomputing works by running information through a series of gates, usually logic gates. A logic gate works as a fork in the road for an electronic circuit. The input will travel one way or another, giving two different outputs. An example logic gate is the AND gate, which has two inputs (A and B) and two different results. If both A and B are 1, the AND gate output will be 1. If only A is 1 and B is 0, the output will be 0 and vice versa. If both A and B are 0, the result will be 0. While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output. In this case, the DNA enters the logic gate as a single or double strand.
If the DNA is double-stranded, the system “digests” the DNA or destroys it, which results in non-fluorescence or “0” output. Conversely, if the DNA is single-stranded, it won’t be digested and instead will be copied by several enzymes in the biocomputing system, resulting in fluorescent RNA or a “1” output. And the output for this type of binary system can be expanded beyond fluorescence or not. For example, a “1” output might be the production of the enzyme insulin, while a “0” may be that no insulin is produced. “This kind of synergy between biology and computation is the essence of biocomputing,” says Stephanie Forrest, a professor and the director of the Biodesign Center for Biocomputing, Security and Society at Arizona State University.
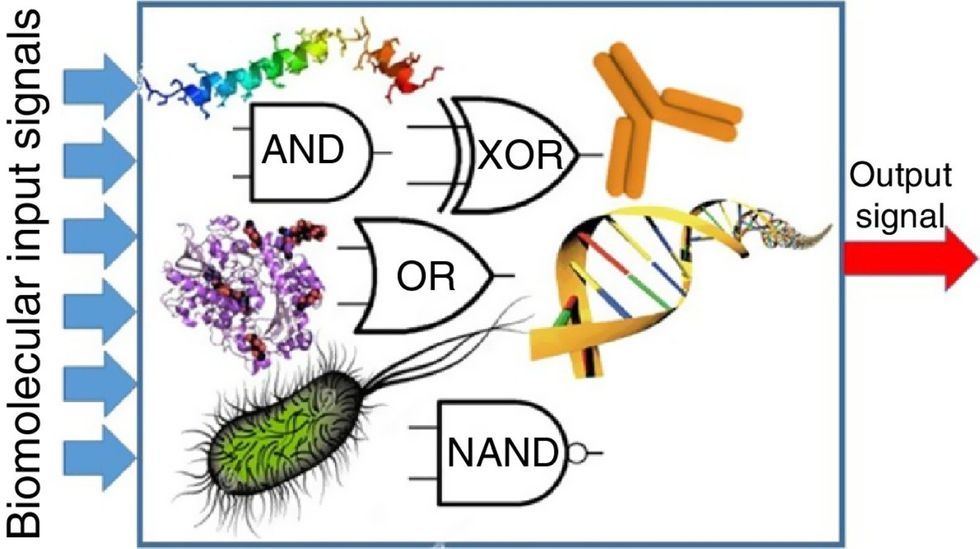
Biocomputing circles are made of DNA, RNA, proteins and even bacteria.
Evgeny Katz
The TRUMPET’s promise
Depending on whether the biocomputing system is placed directly inside a cell within the human body, or run in a test-tube, different environmental factors play a role. When an output is produced inside a cell, the cell's natural processes can amplify this output (for example, a specific protein or DNA strand), creating a solid signal. However, these cells can also be very leaky. “You want the cells to do the thing you ask them to do before they finish whatever their businesses, which is to grow, replicate, metabolize,” Adamala explains. “However, often the gate may be triggered without the right inputs, creating a false positive signal. So that's why natural logic gates are often leaky." While biocomputing outside a cell in a test tube can allow for tighter control over the logic gates, the outputs or signals cannot be amplified by a cell and are less potent.
TRUMPET, which is smaller than a cell, taps into both cellular and non-cellular biocomputing benefits. “At its core, it is a nonliving logic gate system,” Adamala states, “It's a DNA-based logic gate system. But because we use enzymes, and the readout is enzymatic [where an enzyme replicates the fluorescent RNA], we end up with signal amplification." This readout means that the output from the TRUMPET system, a fluorescent RNA strand, can be replicated by nearby enzymes in the platform, making the light signal stronger. "So it combines the best of both worlds,” Adamala adds.
These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body.
The TRUMPET biocomputing process is relatively straightforward. “If the DNA [input] shows up as single-stranded, it will not be digested [by the logic gate], and you get this nice fluorescent output as the RNA is made from the single-stranded DNA, and that's a 1,” Adamala explains. "And if the DNA input is double-stranded, it gets digested by the enzymes in the logic gate, and there is no RNA created from the DNA, so there is no fluorescence, and the output is 0." On the story's leading image above, if the tube is "lit" with a purple color, that is a binary 1 signal for computing. If it's "off" it is a 0.
While still in research, TRUMPET and other biocomputing systems promise significant benefits to personalized healthcare and medicine. These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body. The study’s lead author and graduate student Judee Sharon is already beginning to research TRUMPET's ability for earlier cancer diagnoses. Because the inputs for TRUMPET are single or double-stranded DNA, any mutated or cancerous DNA could theoretically be detected from the platform through the biocomputing process. Theoretically, devices like TRUMPET could be used to detect cancer and other diseases earlier.
Adamala sees TRUMPET not only as a detection system but also as a potential cancer drug delivery system. “Ideally, you would like the drug only to turn on when it senses the presence of a cancer cell. And that's how we use the logic gates, which work in response to inputs like cancerous DNA. Then the output can be the production of a small molecule or the release of a small molecule that can then go and kill what needs killing, in this case, a cancer cell. So we would like to develop applications that use this technology to control the logic gate response of a drug’s delivery to a cell.”
Although platforms like TRUMPET are making progress, a lot more work must be done before they can be used commercially. “The process of translating mechanisms and architecture from biology to computing and vice versa is still an art rather than a science,” says Forrest. “It requires deep computer science and biology knowledge,” she adds. “Some people have compared interdisciplinary science to fusion restaurants—not all combinations are successful, but when they are, the results are remarkable.”
Crickets are low on fat, high on protein, and can be farmed sustainably. They are also crunchy.
In today’s podcast episode, Leaps.org Deputy Editor Lina Zeldovich speaks about the health and ecological benefits of farming crickets for human consumption with Bicky Nguyen, who joins Lina from Vietnam. Bicky and her business partner Nam Dang operate an insect farm named CricketOne. Motivated by the idea of sustainable and healthy protein production, they started their unconventional endeavor a few years ago, despite numerous naysayers who didn’t believe that humans would ever consider munching on bugs.
Yet, making creepy crawlers part of our diet offers many health and planetary advantages. Food production needs to match the rise in global population, estimated to reach 10 billion by 2050. One challenge is that some of our current practices are inefficient, polluting and wasteful. According to nonprofit EarthSave.org, it takes 2,500 gallons of water, 12 pounds of grain, 35 pounds of topsoil and the energy equivalent of one gallon of gasoline to produce one pound of feedlot beef, although exact statistics vary between sources.
Meanwhile, insects are easy to grow, high on protein and low on fat. When roasted with salt, they make crunchy snacks. When chopped up, they transform into delicious pâtes, says Bicky, who invents her own cricket recipes and serves them at industry and public events. Maybe that’s why some research predicts that edible insects market may grow to almost $10 billion by 2030. Tune in for a delectable chat on this alternative and sustainable protein.
Listen on Apple | Listen on Spotify | Listen on Stitcher | Listen on Amazon | Listen on Google
Further reading:
More info on Bicky Nguyen
https://yseali.fulbright.edu.vn/en/faculty/bicky-n...
The environmental footprint of beef production
https://www.earthsave.org/environment.htm
https://www.watercalculator.org/news/articles/beef-king-big-water-footprints/
https://www.frontiersin.org/articles/10.3389/fsufs.2019.00005/full
https://ourworldindata.org/carbon-footprint-food-methane
Insect farming as a source of sustainable protein
https://www.insectgourmet.com/insect-farming-growing-bugs-for-protein/
https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/insect-farming
Cricket flour is taking the world by storm
https://www.cricketflours.com/
https://talk-commerce.com/blog/what-brands-use-cricket-flour-and-why/

Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.