This Revolutionary Medical Breakthrough Is Not a Treatment or a Cure

A doctor examines a patient to determine the cause of her illness.
What is a disease? This seemingly abstract and theoretical question is actually among the most practical questions in all of biomedicine. How patients are diagnosed, treated, managed and excused from various social and moral obligations hinges on the answer that is given. So do issues of how research is done and health care paid for. The question is also becoming one of the most problematic issues that those in health care will face in the next decade.
"The revolution in our understanding of the human genome, molecular biology, and genetics is creating a huge--if little acknowledged--shift in the understanding of what a disease is."
That is because the current conception of disease is undergoing a revolutionary change, fueled by progress in genetics and molecular biology. The consequences of this shift in the definition of disease promise to be as impactful as any other advance in biomedicine has ever been, which is admittedly saying a lot for what is in essence a conceptual change rather than one based on an empirical scientific advance.
For a long time, disease was defined by patient reports of feeling sick. It was not until the twentieth century that a shift occurred away from subjective reports of clusters of symptoms to defining diseases in terms of physiological states. Doctors began to realize that not all symptoms of fever represented the presence of the same disease. Flu got distinguished from malaria. Diseases such as hypertension, osteoporosis, cancer, lipidemia, silent myocardial infarction, retinopathy, blood clots and many others were recognized as not producing any or slight symptoms until suddenly the patient had a stroke or died.
The ability to assess both biology and biochemistry and to predict the consequences of subclinical pathological processes caused a distinction to be made between illness—what a person experiences—and disease—an underlying pathological process with a predictable course. Some conditions, such as Gulf War Syndrome, PTSD, many mental illnesses and fibromyalgia, remain controversial because no underlying pathological process has been found that correlates with them—a landmark criterion for diagnosing disease throughout most of the last century.
"Diseases for which no relationship had ever been posited are being lumped together due to common biochemical causal pathways...that are amenable to the same curative intervention."
The revolution in our understanding of the human genome, molecular biology, and genetics is creating a huge--if little acknowledged--shift in the understanding of what a disease is. A better understanding of the genetic and molecular roots of pathophysiology is leading to the reclassification of many familiar diseases. The test of disease is now not the pathophysiology but the presence of a gene, set of genes or molecular pathway that causes pathophysiology. Just as fever was differentiated into a multitude of diseases in the last century, cancer, cognitive impairment, addiction and many other diseases are being broken or split into many subkinds. And other diseases for which no relationship had ever been posited are being lumped together due to common biochemical causal pathways or the presence of similar dangerous biochemical products that are amenable to the same curative intervention, no matter how disparate the patients' symptoms or organic pathologies might appear.
We used to differentiate ovarian and breast cancers. Now we are thinking of them as outcomes of the same mutations in certain genes in the BRCA regions. They may eventually lump together as BRCA disease.
Other diseases such as familial amyloid polyneuropathy (FAP) which causes polyneuropathy and autonomic dysfunction are being split apart into new types or kinds. The disease is the product of mutations in the transthyretin gene. It was thought to be an autosomal dominant disease with symptomatic onset between 20-40 years of age. However, as genetic testing has improved, it has become clear that FAP's traditional clinical presentation represents a relatively small portion of those with FAP. Many patients with mutations in transthyretin — even mutations commonly seen in traditional FAP patients — do not fit the common clinical presentation. As the mutations begin to be understood, some people that were previously thought to have other polyneuropathies, such as chronic inflammatory demyelinating neuropathy, are now being rediagnosed with newly discovered variants of FAP.
"We are at the start of a major conceptual shift in how we organize the world of disease, and for that matter, health promotion."
Genome-wide association studies are beginning to find many links between diseases not thought to have any connection or association. For example some forms of diabetes, rheumatoid arthritis and thyroid disease may be the products of a small family of genetic mutations.
So why is this shift toward a genetic and molecular diagnostics likely to shake up medicine? One obvious way is that research projects may propose to recruit subjects not according to current standards of disease but on the basis of common genetic mutations or similar errors in biochemical pathways. It won't matter in a future study if subjects in a trial have what today might be termed nicotine addiction or Parkinsonism. If the molecular pathways producing the pathology are the same, then both groups might well wind up in the same trial of a drug.
In addition, what today look like common maladies—pancreatic cancer, severe depression, or acne, for example, could wind up being subdivided into so many highly differentiated versions of these conditions that each must be treated as what we now classify as a rare or ultra-rare disease. Unique biochemical markers or genetic messages may see many diseases broken into a huge number of distinct individual disease entities.
Patients may find that common genetic pathways or multiple effects from a single gene may create new alliances for fund-raising and advocacy. Groups fighting to cure mental and physical illnesses may wind up forgetting about their outward differences in the effort to alter genes or attack common protein markers.
Disease classification appears stable to us—until it isn't. And we are at the start of a major conceptual shift in how we organize the world of disease, and for that matter, health promotion. Classic reductionism, the view that all observable biological phenomena can be explained in terms of underlying chemical and physical principles, may turn out not to be true. But the molecular and genetic revolutions churning through medicine are illustrating that reductionism is going to have an enormous influence on disease classification. That is not a bad thing, but it is something that is going to take a lot to get used to.
DNA- and RNA-based electronic implants may revolutionize healthcare
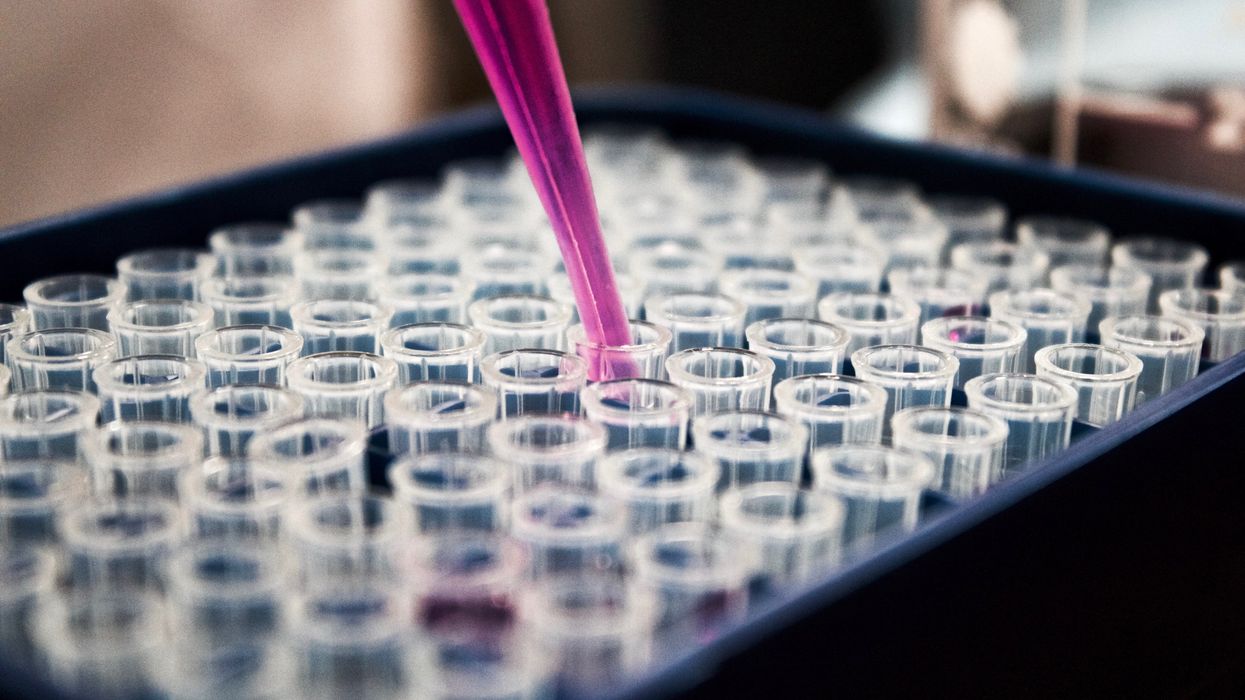
The test tubes contain tiny DNA/enzyme-based circuits, which comprise TRUMPET, a new type of electronic device, smaller than a cell.
Implantable electronic devices can significantly improve patients’ quality of life. A pacemaker can encourage the heart to beat more regularly. A neural implant, usually placed at the back of the skull, can help brain function and encourage higher neural activity. Current research on neural implants finds them helpful to patients with Parkinson’s disease, vision loss, hearing loss, and other nerve damage problems. Several of these implants, such as Elon Musk’s Neuralink, have already been approved by the FDA for human use.
Yet, pacemakers, neural implants, and other such electronic devices are not without problems. They require constant electricity, limited through batteries that need replacements. They also cause scarring. “The problem with doing this with electronics is that scar tissue forms,” explains Kate Adamala, an assistant professor of cell biology at the University of Minnesota Twin Cities. “Anytime you have something hard interacting with something soft [like muscle, skin, or tissue], the soft thing will scar. That's why there are no long-term neural implants right now.” To overcome these challenges, scientists are turning to biocomputing processes that use organic materials like DNA and RNA. Other promised benefits include “diagnostics and possibly therapeutic action, operating as nanorobots in living organisms,” writes Evgeny Katz, a professor of bioelectronics at Clarkson University, in his book DNA- And RNA-Based Computing Systems.
While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output.
Adamala’s research focuses on developing such biocomputing systems using DNA, RNA, proteins, and lipids. Using these molecules in the biocomputing systems allows the latter to be biocompatible with the human body, resulting in a natural healing process. In a recent Nature Communications study, Adamala and her team created a new biocomputing platform called TRUMPET (Transcriptional RNA Universal Multi-Purpose GatE PlaTform) which acts like a DNA-powered computer chip. “These biological systems can heal if you design them correctly,” adds Adamala. “So you can imagine a computer that will eventually heal itself.”
The basics of biocomputing
Biocomputing and regular computing have many similarities. Like regular computing, biocomputing works by running information through a series of gates, usually logic gates. A logic gate works as a fork in the road for an electronic circuit. The input will travel one way or another, giving two different outputs. An example logic gate is the AND gate, which has two inputs (A and B) and two different results. If both A and B are 1, the AND gate output will be 1. If only A is 1 and B is 0, the output will be 0 and vice versa. If both A and B are 0, the result will be 0. While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output. In this case, the DNA enters the logic gate as a single or double strand.
If the DNA is double-stranded, the system “digests” the DNA or destroys it, which results in non-fluorescence or “0” output. Conversely, if the DNA is single-stranded, it won’t be digested and instead will be copied by several enzymes in the biocomputing system, resulting in fluorescent RNA or a “1” output. And the output for this type of binary system can be expanded beyond fluorescence or not. For example, a “1” output might be the production of the enzyme insulin, while a “0” may be that no insulin is produced. “This kind of synergy between biology and computation is the essence of biocomputing,” says Stephanie Forrest, a professor and the director of the Biodesign Center for Biocomputing, Security and Society at Arizona State University.
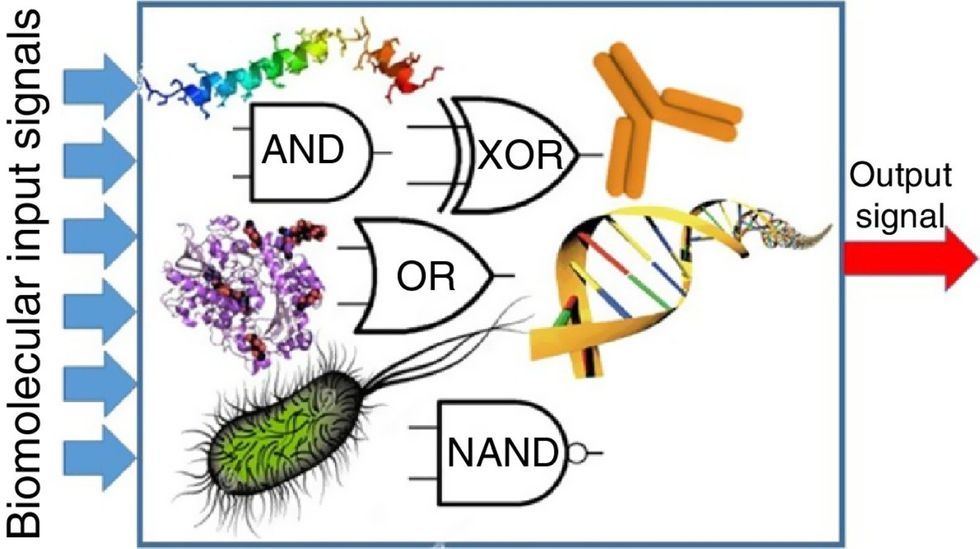
Biocomputing circles are made of DNA, RNA, proteins and even bacteria.
Evgeny Katz
The TRUMPET’s promise
Depending on whether the biocomputing system is placed directly inside a cell within the human body, or run in a test-tube, different environmental factors play a role. When an output is produced inside a cell, the cell's natural processes can amplify this output (for example, a specific protein or DNA strand), creating a solid signal. However, these cells can also be very leaky. “You want the cells to do the thing you ask them to do before they finish whatever their businesses, which is to grow, replicate, metabolize,” Adamala explains. “However, often the gate may be triggered without the right inputs, creating a false positive signal. So that's why natural logic gates are often leaky." While biocomputing outside a cell in a test tube can allow for tighter control over the logic gates, the outputs or signals cannot be amplified by a cell and are less potent.
TRUMPET, which is smaller than a cell, taps into both cellular and non-cellular biocomputing benefits. “At its core, it is a nonliving logic gate system,” Adamala states, “It's a DNA-based logic gate system. But because we use enzymes, and the readout is enzymatic [where an enzyme replicates the fluorescent RNA], we end up with signal amplification." This readout means that the output from the TRUMPET system, a fluorescent RNA strand, can be replicated by nearby enzymes in the platform, making the light signal stronger. "So it combines the best of both worlds,” Adamala adds.
These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body.
The TRUMPET biocomputing process is relatively straightforward. “If the DNA [input] shows up as single-stranded, it will not be digested [by the logic gate], and you get this nice fluorescent output as the RNA is made from the single-stranded DNA, and that's a 1,” Adamala explains. "And if the DNA input is double-stranded, it gets digested by the enzymes in the logic gate, and there is no RNA created from the DNA, so there is no fluorescence, and the output is 0." On the story's leading image above, if the tube is "lit" with a purple color, that is a binary 1 signal for computing. If it's "off" it is a 0.
While still in research, TRUMPET and other biocomputing systems promise significant benefits to personalized healthcare and medicine. These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body. The study’s lead author and graduate student Judee Sharon is already beginning to research TRUMPET's ability for earlier cancer diagnoses. Because the inputs for TRUMPET are single or double-stranded DNA, any mutated or cancerous DNA could theoretically be detected from the platform through the biocomputing process. Theoretically, devices like TRUMPET could be used to detect cancer and other diseases earlier.
Adamala sees TRUMPET not only as a detection system but also as a potential cancer drug delivery system. “Ideally, you would like the drug only to turn on when it senses the presence of a cancer cell. And that's how we use the logic gates, which work in response to inputs like cancerous DNA. Then the output can be the production of a small molecule or the release of a small molecule that can then go and kill what needs killing, in this case, a cancer cell. So we would like to develop applications that use this technology to control the logic gate response of a drug’s delivery to a cell.”
Although platforms like TRUMPET are making progress, a lot more work must be done before they can be used commercially. “The process of translating mechanisms and architecture from biology to computing and vice versa is still an art rather than a science,” says Forrest. “It requires deep computer science and biology knowledge,” she adds. “Some people have compared interdisciplinary science to fusion restaurants—not all combinations are successful, but when they are, the results are remarkable.”
Crickets are low on fat, high on protein, and can be farmed sustainably. They are also crunchy.
In today’s podcast episode, Leaps.org Deputy Editor Lina Zeldovich speaks about the health and ecological benefits of farming crickets for human consumption with Bicky Nguyen, who joins Lina from Vietnam. Bicky and her business partner Nam Dang operate an insect farm named CricketOne. Motivated by the idea of sustainable and healthy protein production, they started their unconventional endeavor a few years ago, despite numerous naysayers who didn’t believe that humans would ever consider munching on bugs.
Yet, making creepy crawlers part of our diet offers many health and planetary advantages. Food production needs to match the rise in global population, estimated to reach 10 billion by 2050. One challenge is that some of our current practices are inefficient, polluting and wasteful. According to nonprofit EarthSave.org, it takes 2,500 gallons of water, 12 pounds of grain, 35 pounds of topsoil and the energy equivalent of one gallon of gasoline to produce one pound of feedlot beef, although exact statistics vary between sources.
Meanwhile, insects are easy to grow, high on protein and low on fat. When roasted with salt, they make crunchy snacks. When chopped up, they transform into delicious pâtes, says Bicky, who invents her own cricket recipes and serves them at industry and public events. Maybe that’s why some research predicts that edible insects market may grow to almost $10 billion by 2030. Tune in for a delectable chat on this alternative and sustainable protein.
Listen on Apple | Listen on Spotify | Listen on Stitcher | Listen on Amazon | Listen on Google
Further reading:
More info on Bicky Nguyen
https://yseali.fulbright.edu.vn/en/faculty/bicky-n...
The environmental footprint of beef production
https://www.earthsave.org/environment.htm
https://www.watercalculator.org/news/articles/beef-king-big-water-footprints/
https://www.frontiersin.org/articles/10.3389/fsufs.2019.00005/full
https://ourworldindata.org/carbon-footprint-food-methane
Insect farming as a source of sustainable protein
https://www.insectgourmet.com/insect-farming-growing-bugs-for-protein/
https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/insect-farming
Cricket flour is taking the world by storm
https://www.cricketflours.com/
https://talk-commerce.com/blog/what-brands-use-cricket-flour-and-why/

Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.