New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
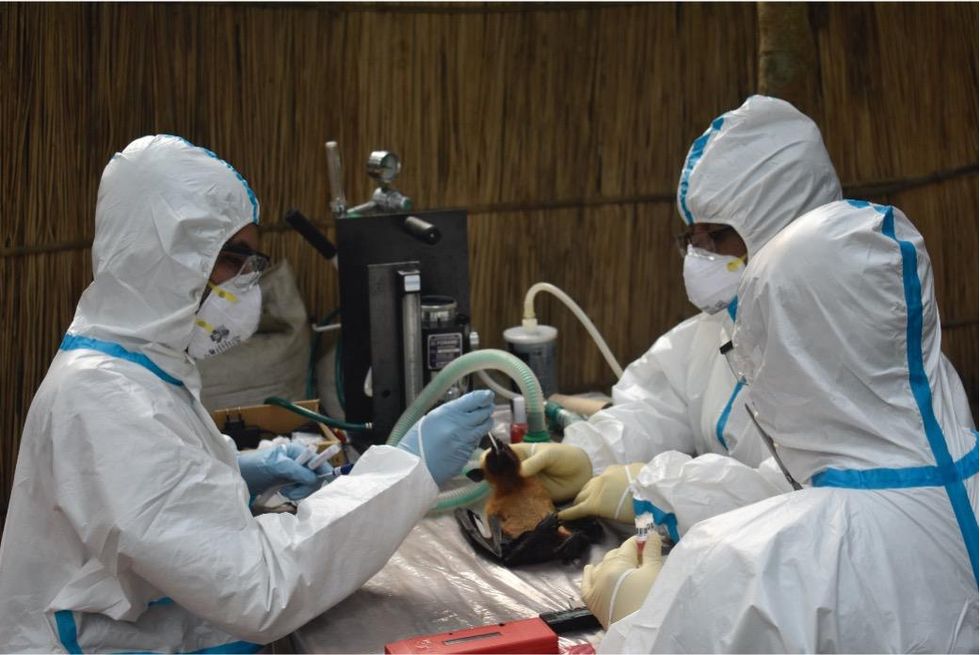
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
Paralyzed By Polio, This British Tea Broker Changed the Course Of Medical History Forever
Robin Cavendish in his special wheelchair with his son Jonathan in the 1960s.
In December 1958, on a vacation with his wife in Kenya, a 28-year-old British tea broker named Robin Cavendish became suddenly ill. Neither he nor his wife Diana knew it at the time, but Robin's illness would change the course of medical history forever.
Robin was rushed to a nearby hospital in Kenya where the medical staff delivered the crushing news: Robin had contracted polio, and the paralysis creeping up his body was almost certainly permanent. The doctors placed Robin on a ventilator through a tracheotomy in his neck, as the paralysis from his polio infection had rendered him unable to breathe on his own – and going off the average life expectancy at the time, they gave him only three months to live. Robin and Diana (who was pregnant at the time with their first child, Jonathan) flew back to England so he could be admitted to a hospital. They mentally prepared to wait out Robin's final days.
But Robin did something unexpected when he returned to the UK – just one of many things that would astonish doctors over the next several years: He survived. Diana gave birth to Jonathan in February 1959 and continued to visit Robin regularly in the hospital with the baby. Despite doctors warning that he would soon succumb to his illness, Robin kept living.
After a year in the hospital, Diana suggested something radical: She wanted Robin to leave the hospital and live at home in South Oxfordshire for as long as he possibly could, with her as his nurse. At the time, this suggestion was unheard of. People like Robin who depended on machinery to keep them breathing had only ever lived inside hospital walls, as the prevailing belief was that the machinery needed to keep them alive was too complicated for laypeople to operate. But Diana and Robin were up for the challenges – and the risks. Because his ventilator ran on electricity, if the house were to unexpectedly lose power, Diana would either need to restore power quickly or hand-pump air into his lungs to keep him alive.
Robin's wheelchair was not only the first of its kind; it became the model for the respiratory wheelchairs that people still use today.
In an interview as an adult, Jonathan Cavendish reflected on his parents' decision to live outside the hospital on a ventilator: "My father's mantra was quality of life," he explained. "He could have stayed in the hospital, but he didn't think that was as good of a life as he could manage. He would rather be two minutes away from death and living a full life."
After a few years of living at home, however, Robin became tired of being confined to his bed. He longed to sit outside, to visit friends, to travel – but had no way of doing so without his ventilator. So together with his friend Teddy Hall, a professor and engineer at Oxford University, the two collaborated in 1962 to create an entirely new invention: a battery-operated wheelchair prototype with a ventilator built in. With this, Robin could now venture outside the house – and soon the Cavendish family became famous for taking vacations. It was something that, by all accounts, had never been done before by someone who was ventilator-dependent. Robin and Hall also designed a van so that the wheelchair could be plugged in and powered during travel. Jonathan Cavendish later recalled a particular family vacation that nearly ended in disaster when the van broke down outside of Barcelona, Spain:
"My poor old uncle [plugged] my father's chair into the wrong socket," Cavendish later recalled, causing the electricity to short. "There was fire and smoke, and both the van and the chair ground to a halt." Johnathan, who was eight or nine at the time, his mother, and his uncle took turns hand-pumping Robin's ventilator by the roadside for the next thirty-six hours, waiting for Professor Hall to arrive in town and repair the van. Rather than being panicked, the Cavendishes managed to turn the vigil into a party. Townspeople came to greet them, bringing food and music, and a local priest even stopped by to give his blessing.
Robin had become a pioneer, showing the world that a person with severe disabilities could still have mobility, access, and a fuller quality of life than anyone had imagined. His mission, along with Hall's, then became gifting this independence to others like himself. Robin and Hall raised money – first from the Ernest Kleinwort Charitable Trust, and then from the British Department of Health – to fund more ventilator chairs, which were then manufactured by Hall's company, Littlemore Scientific Engineering, and given to fellow patients who wanted to live full lives at home. Robin and Hall used themselves as guinea pigs, testing out different models of the chairs and collaborating with scientists to create other devices for those with disabilities. One invention, called the Possum, allowed paraplegics to control things like the telephone and television set with just a nod of the head. Robin's wheelchair was not only the first of its kind; it became the model for the respiratory wheelchairs that people still use today.
Robin went on to enjoy a long and happy life with his family at their house in South Oxfordshire, surrounded by friends who would later attest to his "down-to-earth" personality, his sense of humor, and his "irresistible" charm. When he died peacefully at his home in 1994 at age 64, he was considered the world's oldest-living person who used a ventilator outside the hospital – breaking yet another barrier for what medical science thought was possible.
Kirstie Ennis, an Afghanistan veteran who survived a helicopter crash but lost a limb, pictured in May 2021 at Two Rivers Park in Colorado.
In June 2012, Kirstie Ennis was six months into her second deployment to Afghanistan and recently promoted to sergeant. The helicopter gunner and seven others were three hours into a routine mission of combat resupplies and troop transport when their CH-53D helicopter went down hard.
Miraculously, all eight people onboard survived, but Ennis' injuries were many and severe. She had a torn rotator cuff, torn labrum, crushed cervical discs, facial fractures, deep lacerations and traumatic brain injury. Despite a severely fractured ankle, doctors managed to save her foot, for a while at least.
In November 2015, after three years of constant pain and too many surgeries to count, Ennis relented. She elected to undergo a lower leg amputation but only after she completed the 1,000-mile, 72-day Walking with the Wounded journey across the UK.
On Veteran's Day of that year, on the other side of the country, orthopedic surgeon Cato Laurencin announced a moonshot challenge he was setting out to achieve on behalf of wounded warriors like Ennis: the Hartford Engineering A Limb (HEAL) Project.
Laurencin, who is a University of Connecticut professor of chemical, materials and biomedical engineering, teamed up with experts in tissue bioengineering and regenerative medicine from Harvard, Columbia, UC Irvine and SASTRA University in India. Laurencin and his colleagues at the Connecticut Convergence Institute for Translation in Regenerative Engineering made a bold commitment to regenerate an entire limb within 15 years – by the year 2030.

Dr. Cato Laurencin pictured in his office at UConn.
Photo Credit: UConn
Regenerative Engineering -- A Whole New Field
Limb regeneration in humans has been a medical and scientific fascination for decades, with little to show for the effort. However, Laurencin believes that if we are to reach the next level of 21st century medical advances, this puzzle must be solved.
An estimated 185,000 people undergo upper or lower limb amputation every year. Despite the significant advances in electromechanical prosthetics, these individuals still lack the ability to perform complex functions such as sensation for tactile input, normal gait and movement feedback. As far as Laurencin is concerned, the only clinical answer that makes sense is to regenerate a whole functional limb.
Laurencin feels other regeneration efforts were hampered by their siloed research methods with chemists, surgeons, engineers all working separately. Success, he argues, requires a paradigm shift to a trans-disciplinary approach that brings together cutting-edge technologies from disparate fields such as biology, material sciences, physical, chemical and engineering sciences.
As the only surgeon ever inducted into the academies of Science, Medicine and Innovation, Laurencin is uniquely suited for the challenge. He is regarded as the founder of Regenerative Engineering, defined as the convergence of advanced materials sciences, stem cell sciences, physics, developmental biology and clinical translation for the regeneration of complex tissues and organ systems.
But none of this is achievable without early clinician participation across scientific fields to develop new technologies and a deeper understanding of how to harness the body's innate regenerative capabilities. "When I perform a surgical procedure or something is torn or needs to be repaired, I count on the body being involved in regenerating tissue," he says. "So, understanding how the body works to regenerate itself and harnessing that ability is an important factor for the regeneration process."
The Birth of the Vision
Laurencin's passion for regeneration began when he was a sports medicine fellow at Cornell University Medical Center in the early 1990s. There he saw a significant number of injuries to the anterior cruciate ligament (ACL), the major ligament that stabilizes the knee. He believed he could develop a better way to address those injuries using biomaterials to regenerate the ligament. He sketched out a preliminary drawing on a napkin one night over dinner. He has spent the next 30 years regenerating tissues, including the patented L-C ligament.
As chair of Orthopaedic Surgery at the University of Virginia during the peak of the wars in Iraq and Afghanistan, Laurencin treated military personnel who survived because of improved helmets, body armor and battlefield medicine but were left with more devastating injuries, including traumatic brain injuries and limb loss.
"I was so honored to care for them and I so admired their steadfast courage that I became determined to do something big for them," says Laurencin.
When he tells people about his plans to regrow a limb, he gets a lot of eye rolls, which he finds amusing but not discouraging. Growing bone cells was relatively new when he was first focused on regenerating bone in 1987 at MIT; in 2007 he was well on his way to regenerating ligaments at UVA when many still doubted that ligaments could even be reconstructed. He and his team have already regenerated torn rotator cuff tendons and ACL ligaments using a nano-textured fabric seeded with stem cells.
Even as a finalist for the $4 million NIH Pioneer Award for high-risk/high-reward research, he faced a skeptical scientific audience in 2014. "They said, 'Well what do you plan to do?' I said 'I plan to regenerate a whole limb in people.' There was a lot of incredulousness. They stared at me and asked a lot of questions. About three days later, I received probably the best score I've ever gotten on an NIH grant."
In the Thick of the Science
Humans are born with regenerative abilities--two-year-olds have regrown fingertips--but lose that ability with age. Salamanders are the only vertebrates that can regenerate lost body parts as adults; axolotl, the rare Mexican salamander, can grow extra limbs.
The axolotl is important as a model organism because it is a four-footed vertebrate with a similar body plan to humans. Mapping the axolotl genome in 2018 enhanced scientists' genetic understanding of their evolution, development, and regeneration. Being easy to breed in captivity allowed the HEAL team to closely study these amphibians and discover a new cell type they believe may shed light on how to mimic the process in humans.
"Whenever limb regeneration takes place in the salamander, there is a huge amount of something called heparan sulfate around that area," explains Laurencin. "We thought, 'What if this heparan sulfate is the key ingredient to allowing regeneration to take place?' We found these groups of cells that were interspersed in tissues during the time of regeneration that seemed to have connections to each other that expressed this heparan sulfate."
Called GRID (Groups that are Regenerative, Interspersed and Dendritic), these cells were also recently discovered in mice. While GRID cells don't regenerate as well in mice as in salamanders, finding them in mammals was significant.
"If they're found in mice. we might be able to find these in humans in some form," Laurencin says. "We think maybe it will help us figure out regeneration or we can create cells that mimic what grid cells do and create an artificial grid cell."
What Comes Next?
Laurencin and his team have individually engineered and made every single tissue in the lower limb, including bone, cartilage, ligament, skin, nerve, blood vessels. Regenerating joints and joint tissue is the next big mile marker, which Laurencin sees as essential to regenerating a limb that functions and performs in the way he envisions.
"Using stem cells and amnion tissue, we can regenerate joints that are damaged, and have severe arthritis," he says. "We're making progress on all fronts, and making discoveries we believe are going to be helping people along the way."
That focus and advancement is vital to Ennis. After laboring over the decision to have her leg amputated below the knee, she contracted MRSA two weeks post-surgery. In less than a month, she went from a below-the-knee-amputee to a through-the-knee amputee to an above-the-knee amputee.
"A below-the-knee amputation is night-and-day from above-the-knee," she said. "You have to relearn everything. You're basically a toddler."
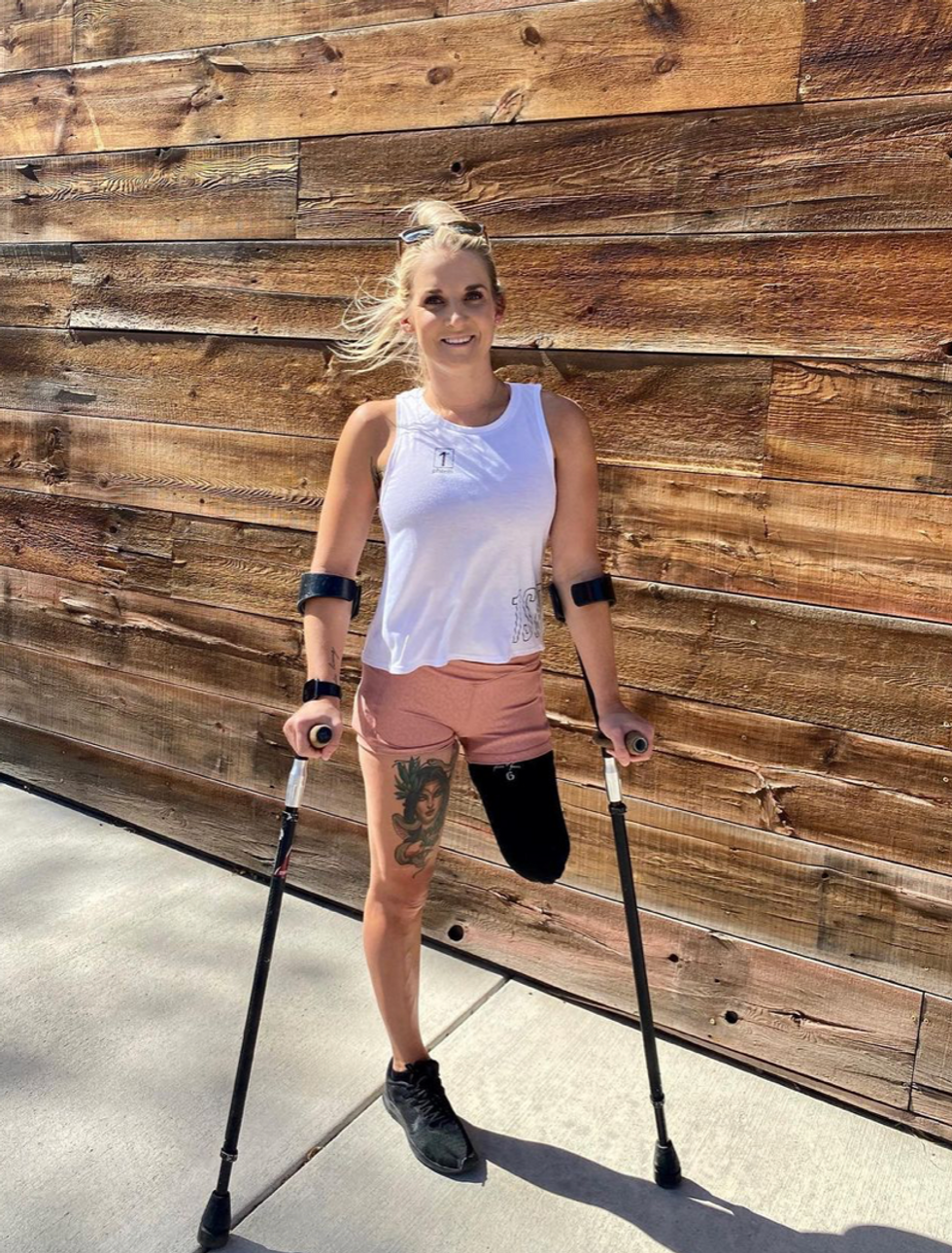
Kirstie Ennis pictured in July 2020.
Photo Credit: Ennis' Instagram
The clock is ticking on the timeline Laurencin set for himself. Nine years might seem like forever if you're doing time but it might appear fleeting when you're trying to create something that's never been done before. But Laurencin isn't worried. He's convinced time is on his side.
"Every week, I receive an email or a call from someone, maybe a mother whose child has lost a finger or I'm in communication with a disabled American veteran who wants to know how the progress is going. That energizes me to continue to work hard to try to create these sorts of solutions because we're talking about people and their lives."
He devotes about 60 hours a week to the project and the roughly 100 students, faculty and staff who make up the HEAL team at the Convergence Institute seem acutely aware of what's at stake and appear equally dedicated.
"We're in the thick of the science in terms of making this happen," says Laurencin. "We've moved from making the impossible possible to making the possible a reality. That's what science is all about."

