New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
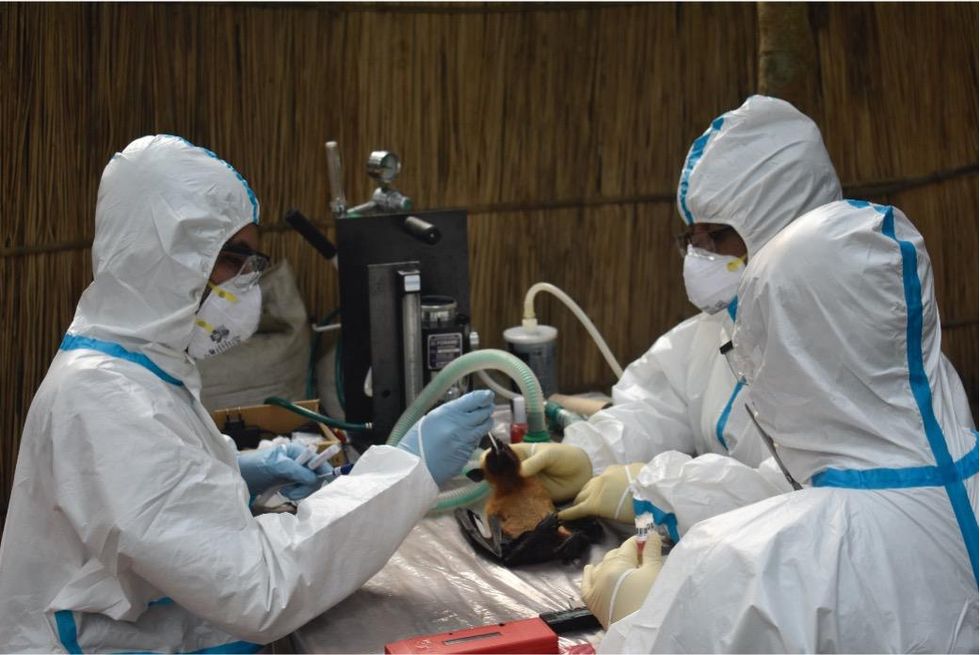
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
How Emerging Technologies Can Help Us Fight the New Coronavirus
A security officer in a protective mask checks the temperature of a passenger following the outbreak of a new coronavirus, at an expressway toll station on the eve of the Chinese Lunar New Year celebrations, in Xianning.
In nature, few species remain dominant for long. Any sizable population of similar individuals offers immense resources to whichever parasite can evade its defenses, spreading rapidly from one member to the next.
Which will prove greater: our defenses or our vulnerabilities?
Humans are one such dominant species. That wasn't always the case: our hunter-gatherer ancestors lived in groups too small and poorly connected to spread pathogens like wildfire. Our collective vulnerability to pandemics began with the dawn of cities and trade networks thousands of years ago. Roman cities were always demographic sinks, but never more so than when a pandemic agent swept through. The plague of Cyprian, the Antonine plague, the plague of Justinian – each is thought to have killed over ten million people, an appallingly high fraction of the total population of the empire.
With the advent of sanitation, hygiene, and quarantines, we developed our first non-immunological defenses to curtail the spread of plagues. With antibiotics, we began to turn the weapons of microbes against our microbial foes. Most potent of all, we use vaccines to train our immune systems to fight pathogens before we are even exposed. Edward Jenner's original vaccine alone is estimated to have saved half a billion lives.
It's been over a century since we suffered from a swift and deadly pandemic. Even the last deadly influenza of 1918 killed only a few percent of humanity – nothing so bad as any of the Roman plagues, let alone the Black Death of medieval times.
How much of our recent winning streak has been due to luck?
Much rides on that question, because the same factors that first made our ancestors vulnerable are now ubiquitous. Our cities are far larger than those of ancient times. They're inhabited by an ever-growing fraction of humanity, and are increasingly closely connected: we now routinely travel around the world in the course of a day. Despite urbanization, global population growth has increased contact with wild animals, creating more opportunities for zoonotic pathogens to jump species. Which will prove greater: our defenses or our vulnerabilities?
The tragic emergence of coronavirus 2019-nCoV in Wuhan may provide a test case. How devastating this virus will become is highly uncertain at the time of writing, but its rapid spread to many countries is deeply worrisome. That it seems to kill only the already infirm and spare the healthy is small comfort, and may counterintuitively assist its spread: it's easy to implement a quarantine when everyone infected becomes extremely ill, but if carriers may not exhibit symptoms as has been reported, it becomes exceedingly difficult to limit transmission. The virus, a distant relative of the more lethal SARS virus that killed 800 people in 2002 to 2003, has evolved to be transmitted between humans and spread to 18 countries in just six weeks.
Humanity's response has been faster than ever, if not fast enough. To its immense credit, China swiftly shared information, organized and built new treatment centers, closed schools, and established quarantines. The Coalition for Epidemic Preparedness Innovations, which was founded in 2017, quickly funded three different companies to develop three different varieties of vaccine: a standard protein vaccine, a DNA vaccine, and an RNA vaccine, with more planned. One of the agreements was signed after just four days of discussion, far faster than has ever been done before.
The new vaccine candidates will likely be ready for clinical trials by early summer, but even if successful, it will be additional months before the vaccine will be widely available. The delay may well be shorter than ever before thanks to advances in manufacturing and logistics, but a delay it will be.
The 1918 influenza virus killed more than half of its victims in the United Kingdom over just three months.
If we faced a truly nasty virus, something that spreads like pandemic influenza – let alone measles – yet with the higher fatality rate of, say, H7N9 avian influenza, the situation would be grim. We are profoundly unprepared, on many different levels.
So what would it take to provide us with a robust defense against pandemics?
Minimize the attack surface: 2019-nCoV jumped from an animal, most probably a bat, to humans. China has now banned the wildlife trade in response to the epidemic. Keeping it banned would be prudent, but won't be possible in all nations. Still, there are other methods of protection. Influenza viruses commonly jump from birds to pigs to humans; the new coronavirus may have similarly passed through a livestock animal. Thanks to CRISPR, we can now edit the genomes of most livestock. If we made them immune to known viruses, and introduced those engineered traits to domesticated animals everywhere, we would create a firewall in those intermediate hosts. We might even consider heritably immunizing the wild organisms most likely to serve as reservoirs of disease.
None of these defenses will be cheap, but they'll be worth every penny.
Rapid diagnostics: We need a reliable method of detection costing just pennies to be available worldwide inside of a week of discovering a new virus. This may eventually be possible thanks to a technology called SHERLOCK, which is based on a CRISPR system more commonly used for precision genome editing. Instead of using CRISPR to find and edit a particular genome sequence in a cell, SHERLOCK programs it to search for a desired target and initiate an easily detected chain reaction upon discovery. The technology is capable of fantastic sensitivity: with an attomolar (10-18) detection limit, it senses single molecules of a unique DNA or RNA fingerprint, and the components can be freeze-dried onto paper strips.
Better preparations: China acted swiftly to curtail the spread of the Wuhan virus with traditional public health measures, but not everything went as smoothly as it might have. Most cities and nations have never conducted a pandemic preparedness drill. Best give people a chance to practice keeping the city barely functional while minimizing potential exposure events before facing the real thing.
Faster vaccines: Three months to clinical trials is too long. We need a robust vaccine discovery and production system that can generate six candidates within a week of the pathogen's identification, manufacture a million doses the week after, and scale up to a hundred million inside of a month. That may be possible for novel DNA and RNA-based vaccines, and indeed anything that can be delivered using a standardized gene therapy vector. For example, instead of teaching each person's immune system to evolve protective antibodies by showing it pieces of the virus, we can program cells to directly produce known antibodies via gene therapy. Those antibodies could be discovered by sifting existing diverse libraries of hundreds of millions of candidates, computationally designed from scratch, evolved using synthetic laboratory ecosystems, or even harvested from the first patients to report symptoms. Such a vaccine might be discovered and produced fast enough at scale to halt almost any natural pandemic.
Robust production and delivery: Our defenses must not be vulnerable to the social and economic disruptions caused by a pandemic. Unfortunately, our economy selects for speed and efficiency at the expense of robustness. Just-in-time supply chains that wing their way around the world require every node to be intact. If workers aren't on the job producing a critical component, the whole chain breaks until a substitute can be found. A truly nasty pandemic would disrupt economies all over the world, so we will need to pay extra to preserve the capacity for independent vertically integrated production chains in multiple nations. Similarly, vaccines are only useful if people receive them, so delivery systems should be as robustly automated as possible.
None of these defenses will be cheap, but they'll be worth every penny. Our nations collectively spend trillions on defense against one another, but only billions to protect humanity from pandemic viruses known to have killed more people than any human weapon. That's foolish – especially since natural animal diseases that jump the species barrier aren't the only pandemic threats.
We will eventually make our society immune to naturally occurring pandemics, but that day has not yet come, and future pandemic viruses may not be natural.
The complete genomes of all historical pandemic viruses ever to have been sequenced are freely available to anyone with an internet connection. True, these are all agents we've faced before, so we have a pre-existing armory of pharmaceuticals and vaccines and experience. There's no guarantee that they would become pandemics again; for example, a large fraction of humanity is almost certainly immune to the 1918 influenza virus due to exposure to the related 2009 pandemic, making it highly unlikely that the virus would take off if released.
Still, making the blueprints publicly available means that a large and growing number of people with the relevant technical skills can single-handedly make deadly biological agents that might be able to spread autonomously -- at least if they can get their hands on the relevant DNA. At present, such people most certainly can, so long as they bother to check the publicly available list of which gene synthesis companies do the right thing and screen orders -- and by implication, which ones don't.
One would hope that at least some of the companies that don't advertise that they screen are "honeypots" paid by intelligence agencies to catch would-be bioterrorists, but even if most of them are, it's still foolish to let individuals access that kind of destructive power. We will eventually make our society immune to naturally occurring pandemics, but that day has not yet come, and future pandemic viruses may not be natural. Hence, we should build a secure and adaptive system capable of screening all DNA synthesis for known and potential future pandemic agents... without disclosing what we think is a credible bioweapon.
Whether or not it becomes a global pandemic, the emergence of Wuhan coronavirus has underscored the need for coordinated action to prevent the spread of pandemic disease. Let's ensure that our reactive response minimally prepares us for future threats, for one day, reacting may not be enough.
This Dog's Nose Is So Good at Smelling Cancer That Scientists Are Trying to Build One Just Like It
Claire Guest, co-founder of Medical Detection Dogs, with Daisy, whom she credits with saving her life.
Daisy wouldn't leave Claire Guest alone. Instead of joining Guest's other dogs for a run in the park, the golden retriever with the soulful eyes kept nudging Guest's chest, and stared at her intently, somehow hoping she'd get the message.
"I was incredibly lucky to be told by Daisy."
When Guest got home, she detected a tiny lump in one of her breasts. She dismissed it, but her sister, who is a family doctor, insisted she get it checked out.
That saved her life. A series of tests, including a biopsy and a mammogram, revealed the cyst was benign. But doctors discovered a tumor hidden deep inside her chest wall, an insidious malignancy that normally isn't detected until the cancer has rampaged out of control throughout the body. "My prognosis would have been very poor," says Guest, who is an animal behavioralist. "I was incredibly lucky to be told by Daisy."
Ironically, at the time, Guest was training hearing dogs for the deaf—alerting them to doorbells or phones--for a charitable foundation. But she had been working on a side project to harness dogs' exquisitely sensitive sense of smell to spot cancer at its earliest and most treatable stages. When Guest was diagnosed with cancer two decades ago, however, the use of dogs to detect diseases was in its infancy and scientific evidence was largely anecdotal.
In the years since, Guest and the British charitable foundation she co-founded with Dr. John Church in 2008, Medical Detection Dogs (MDD), has shown that dogs can be trained to detect odors that predict a looming medical crisis hours in advance, in the case of diabetes or epilepsy, as well as the presence of cancers.
In a proof of principle study published in the BMJ in 2004, they showed dogs had better than a 40 percent success rate in identifying bladder cancer, which was significantly better than random chance (14 percent). Subsequent research indicated dogs can detect odors down to parts per trillion, which is the equivalent of sniffing out a teaspoon of sugar in two Olympic size swimming pools (a million gallons).
American scientists are devising artificial noses that mimic dogs' sense of smell, so these potentially life-saving diagnostic tools are widely available.
But the problem is "dogs can't be scaled up"—it costs upwards of $25,000 to train them—"and you can't keep a trained dog in every oncology practice," says Guest.
The good news is that the pivotal 2004 BMJ paper caught the attention of two American scientists—Andreas Mershin, a physicist at MIT, and Wen-Yee Yee, a chemistry professor at The University of Texas at El Paso. They have joined Guest's quest to leverage canines' highly attuned olfactory systems and devise artificial noses that mimic dogs' sense of smell, so these potentially life-saving diagnostic tools are widely available.
"What we do know is that this is real," says Guest. "Anything that can improve diagnosis of cancer is something we ought to know about."
Dogs have routinely been used for centuries as trackers for hunting and more recently, for ferreting out bombs and bodies. Dogs like Daisy, who went on to become a star performer in Guest's pack of highly trained cancer detecting canines before her death in 2018, have shared a special bond with their human companions for thousands of years. But their vastly superior olfaction is the result of simple anatomy.
Humans possess about six million olfactory receptors—the antenna-like structures inside cell membranes in our nose that latch on to the molecules in the air when we inhale. In contrast, dogs have about 300 million of them and the brain region that analyzes smells is, proportionally, about 40 times greater than ours.
Research indicates that cancerous cells interfere with normal metabolic processes, prompting them to produce volatile organic compounds (VOCs), which enter the blood stream and are either exhaled in our breath or excreted in urine. Dogs can identify these VOCs in urine samples at the tiniest concentrations, 0.001 parts per million, and can be trained to identify the specific "odor fingerprint" of different cancers, although teaching them how to distinguish these signals from background odors is far more complicated than training them to detect drugs or explosives.
For the past fifteen years, Andreas Mershin of MIT has been grappling with this complexity in his quest to devise an artificial nose, which he calls the Nano-Nose, first as a military tool to spot land mines and IEDS, and more recently as a cancer detection tool that can be used in doctors' offices. The ultimate goal is to create an easy-to-use olfaction system powered by artificial intelligence that can fit inside of smartphones and can replicate dogs' ability to sniff out early signs of prostate cancer, which could eliminate a lot of painful and costly biopsies.
Trained canines have a better than 90 percent accuracy in spotting prostate cancer, which is normally difficult to detect. The current diagnostic, the prostate specific antigen test, which measures levels of certain immune system cells associated with prostate cancer, has about as much accuracy "as a coin toss," according to the scientist who discovered PSA. These false positives can lead to unnecessary and horrifically invasive biopsies to retrieve tissue samples.
So far, Mershin's prototype device has the same sensitivity as the dogs—and can detect odors at parts per trillion—but it still can't distinguish that cancer smell in individual human patients the way a dog can. "What we're trying to understand from the dogs is how they look at the data they are collecting so we can copy it," says Mershin. "We still have to make it intelligent enough to know what it is looking at—what we are lacking is artificial dog intelligence."

The intricate parts of the artificial nose are designed to fit inside a smartphone.
At UT El Paso, Wen-Yee Lee and her research team has used the canine olfactory system as a model for a new screening test for prostate cancer, which has a 92 percent accuracy in tests of urine samples and could be eventually developed as a kit similar to the home pregnancy test. "If dogs can do it, we can do it better," says Lee, whose husband was diagnosed with prostate cancer in 2005.
The UT scientists used samples from about 150 patients, and looked at about 9,000 compounds before they were able to zero in on the key VOCs that are released by prostate cancers—"it was like finding a needle in the haystack," says Lee. But a more reliable test that can also distinguish which cancers are more aggressive could help patients decide their best treatment options and avoid invasive procedures that can render them incontinent and impotent.
"This is much more accurate than the PSA—we were able to see a very distinct difference between people with prostate cancer and those without cancer," says Lee, who has been sharing her research with Guest and hopes to have the test on the market within the next few years.
In the meantime, Guest's foundation has drawn the approving attention of royal animal lovers: Camilla, the Duchess of Cornwall, is a patron, which opened up the charitable floodgates and helped legitimize MDD in the scientific community. Even Camilla's mother-in-law, Queen Elizabeth, has had a demonstration of these canny canines' unique abilities.

Claire Guest, and two of MDDs medical detection dogs, Jodie and Nimbus, meet with queen Elizabeth.
"She actually held one of my [artificial] noses in her hand and asked really good questions, including things we hadn't thought of, like the range of how far away a dog can pick up the scent or if this can be used to screen for malaria," says Mershin. "I was floored by this curious 93-year-old lady. Half of humanity's deaths are from chronic diseases and what the dogs are showing is a whole new way of understanding holistic diseases of the system."


