New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
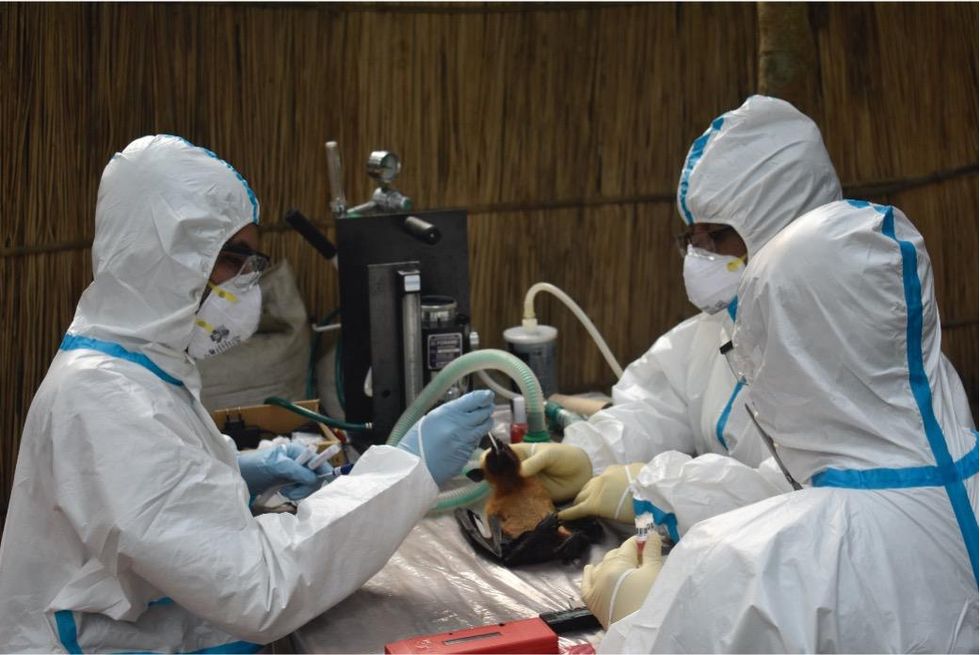
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
In China, Prisoners of Conscience Are Being Murdered for Their Organs to Fuel Transplant Tourism
A somber photo of the Great Wall of China.
Organ transplantation can dramatically improve or save lives. A heart transplant can literally give a person a new lease of life, while a kidney transplant frees the recipient from lengthy spells on dialysis.
A people's tribunal in London has recently found that in China, organs are sourced from prisoners of conscience who are killed on demand to fuel the lucrative organ transplantation market.
To protect the integrity of organ transplantation, there are strict ethical guidelines. When organs are sourced from deceased persons, the donation must be voluntary, donors must die naturally before any organs are taken, and death must not be hastened to provide organs. These ethical guidelines protect donors and provide assurance to transplant recipients that their organs have been sourced ethically.
However, not all countries follow these ethical guidelines. A people's tribunal in London has recently found that in China, organs are sourced from prisoners of conscience who are killed on demand to fuel the lucrative organ transplantation market. This conclusion, reported at the United Nations Human Rights Council on September 24, was not reached lightly.
The independent China Tribunal, made up of four human rights lawyers, one surgeon with transplant experience, an academic who specialises in China studies and a businessman with human rights interests, spent over a year looking at written materials and heard evidence from over 50 witnesses in five days of hearings. Their grim conclusion, that prisoners of conscience are murdered for their organs, confirms the findings of earlier investigations.
Questions first arose over China's transplant system when the numbers of transplants rose dramatically after 2000. Transplant capacity rapidly increased; new infrastructure was built and staff were trained. Hospital websites offered livers, hearts and kidneys available in a matter of days or weeks, for a price. Foreigners were encouraged to come to China to avoid lengthy transplant waiting lists in their home countries.
At the time, it was a mystery as to how China had a ready supply of organs, despite having no volunteer donation system. Eventually, in 2006, the Chinese government stated that organs were removed from prisoners who had been found guilty and sentenced to the death penalty. But this explanation did not ring true. Death row prisoners often have poor health, including high rates of infectious diseases, making them poor candidates for donation. By contrast, the organs offered for sale were promised to be healthy.
In 2006, the first clues about the source of the organs emerged. A woman called Annie reported that her surgeon husband had been present during organ removal from Falun Gong practitioners who were still breathing as the scalpels cut into them. A subsequent investigation by two Canadian human rights lawyers examined multiple sources of evidence, concluding that murdered Falun Gong practitioners were indeed the source of the organs.
The evidence included testimony from practitioners who had been imprisoned, tortured, and later released. During imprisonment, many practitioners reported blood and other medical tests examining the health of their organs—tests that were not performed on any other prisoners. Phone calls made to Chinese hospitals by investigators posing as patients were offered rapid access to fresh organs from Falun Gong practitioners. The organs were guaranteed to be healthy, as the practice forbids smoking tobacco and drinking alcohol.
Since 2006, evidence has continued to accumulate. China has a huge transplant industry and no plausible source of voluntary organ donations. Unlike the rest of the world, Chinese waiting times remain very short. Foreigners continue to come to China to avoid lengthy waiting lists. Prisoners of conscience, including Tibetans and Uyghurs as well as Falun Gong practitioners, are still being imprisoned and medically tested.
The Chinese government continues to deny these crimes, claiming that there is a volunteer donor system in place.
The China Tribunal heard from Uyghur witnesses who had recently been inside the notorious labour camps (also called "re-education" centers) in Xin Xiang. The witnesses reported terrible conditions, including overcrowding and torture, and were forced to have medical examinations. They saw other prisoners disappear without explanation following similar medical tests. As recently as 2018, doctors in Chinese hospitals were promising potential patients healthy Falun Gong organs in taped phone calls.
The Chinese government continues to deny these crimes, claiming that there is a volunteer donor system in place. In the Chinese system, prisoners are counted as volunteers.
China's forced organ harvesting from prisoners of conscience has international implications. A recent study found that most published Chinese transplant research is based on organs sourced from prisoners. International ethical guidance prohibits taking organs from prisoners and prohibits publication of research based on transplanted material from prisoners. The authors of that study called for retractions of the papers, some of which are in well-known scientific journals. So far Transplantation and PLOS One are among the journals that have already retracted over twenty articles in response. On questioning from the editors, the authors of the papers failed to respond or could not verify that the organs in the transplant research came from volunteers.
The international community has a moral obligation to act together to stop forced organ harvesting in China.
The China Tribunal concluded that forced organ harvesting remains China's main source of transplant organs. In their view, the commission of Crimes Against Humanity against the Uyghurs and Falun Gong has been proved beyond reasonable doubt. By their actions, the Chinese government has turned a life-saving altruistic practice into our worst nightmare. The international community has a moral obligation to act together to stop forced organ harvesting in China, and end these crimes against humanity.
Taylor Schreiber, now 39, runs an immunotherapy company testing drugs that may be less toxic alternative to chemotherapy.
It looked like only good things were ahead of Taylor Schreiber in 2010.
Schreiber had just finished his PhD in cancer biology and was preparing to return to medical school to complete his degree. He also had been married a year, and, like any young newlyweds up for adventure, he and his wife Nicki decided to go backpacking in the Costa Rican rainforest.
He was 31, and it was April Fool's Day—but no joke.
During the trip, he experienced a series of night sweats and didn't think too much about it. Schreiber hadn't been feeling right for a few weeks and assumed he had a respiratory infection. Besides, they were sleeping outdoors in a hot, tropical jungle.
But the night sweats continued even after he got home, leaving his mattress so soaked in the morning it was if a bucket of water had been dumped on him overnight. On instinct, he called one of his thesis advisors at the Sylvester Comprehensive Cancer Center in Florida and described his symptoms.
Dr. Joseph Rosenblatt didn't hesitate. "It sounds like Hodgkins. Come see me tomorrow," he said.
The next day, Schreiber was diagnosed with Stage 3b Hodgkin Lymphoma, which meant the disease was advanced. He was 31, and it was April Fool's Day—but no joke.
"I was scared to death," he recalls. "[Thank] goodness it's one of those cancers that is highly treatable. But being 31 years old and all of a sudden being told that you have a 30 percent of mortality within the next two years wasn't anything that I was relieved about."
For Schreiber, the diagnosis was a personal and professional game-changer. He couldn't work in the hospital as a medical student while undergoing chemotherapy, so he wound up remaining in his post-doctorate lab for another two years. The experience also solidified his decision to apply his scientific and medical knowledge to drug development.
Today, now 39, Schreiber is co-founder, director and chief scientific officer of Shattuck Labs, an immuno-oncology startup, and the developer of several important research breakthroughs in the field of immunotherapy.
After his diagnosis, he continued working full-time as a postdoc, while undergoing an aggressive chemotherapy regimen.
"These days, I look back on [my cancer] and think it was one of the luckiest things that ever happened to me," he says. "In medical school, you learn what it is to treat people and learn about the disease. But there is nothing like being a patient to teach you another side of medicine."
Medicine first called to Schreiber when his maternal grandfather was dying from lung cancer complications. Schreiber's uncle, a radiologist at the medical center where his grandfather was being treated, took him on a tour of his department and showed him images of the insides of his body on an ultrasound machine.
Schreiber was mesmerized. His mother was a teacher and his dad sold windows, so medicine was not something to which he had been routinely exposed.
"This weird device was like looking through jelly, and I thought that was the coolest thing ever," he says.
The experience led him to his first real job at the Catholic Medical Center in Manchester, NH, then to a semester-long internship program during his senior year in high school in Concord Hospital's radiology department.
"This was a great experience, but it also made clear that there was not any meaningful way to learn or contribute to medicine before you obtained a medical degree," says Schreiber, who enrolled in Bucknell College to study biology.
Bench science appealed to him, and he volunteered in Dr. Jing Zhou's nephrology department lab at the Harvard Institutes of Medicine. Under the mentorship of one of her post-docs, Lei Guo, he learned a range of critical techniques in molecular biology, leading to their discovery of a new gene related to human polycystic kidney disease and his first published paper.
Before his cancer diagnosis, Schreiber also volunteered in the lab of Dr. Robert "Doc" Sackstein, a world-renowned bone marrow transplant physician and biomedical researcher, and his interests began to shift towards immunology.
"He was just one of those dynamic people who has a real knack for teaching, first of all, and for inspiring people to want to learn more and ask hard questions and understand experimental medicine," Schreiber says.
It was there that he learned the scientific method and the importance of incorporating the right controls in experiments—a simple idea, but difficult to perform well. He also made what Sackstein calls "a startling discovery" about chemokines, which are signaling proteins that can activate an immune response.
As immune cells travel around our bodies looking for potential sources of infection or disease, they latch onto blood vessel walls and "sniff around" for specific chemical cues that indicate a source of infection. Schreiber and his colleagues designed a system that mimics the blood vessel wall, allowing them to define which chemical cues efficiently drive immune cell migration from the blood into tissues.
Schreiber received the best overall research award in 2008 from the National Student Research Foundation. But even as Schreiber's expertise about immunology grew, his own immune system was about to fight its hardest battle.
After his diagnosis, he continued working full-time as a postdoc in the lab of Eckhard Podack, then chair of the microbiology and immunology department at the University of Miami's Leonard M. Miller School of Medicine.
At the same time, Schreiber began an aggressive intravenous chemotherapy regimen of adriamycin, bleomycin, vincristine and dacarbazine, every two weeks, for 6 months. His wife Nicki, an obgyn, transferred her residency from Emory University in Atlanta to Miami so they could be together.
"It was a weird period. I mean, it made me feel good to keep doing things and not just lay idle," he said. "But by the second cycle of chemo, I was immunosuppressed and losing my hair and wore a face mask walking around the lab, which I was certainly self-conscious. But everyone around me didn't make me feel like an alien so I just went about my business."
The experience reinforced his desire to stay in immunology, especially after having taken the most toxic chemotherapies.
He stayed home the day after chemo when he felt his worst, then rested his body and timed exercise to give the drugs the best shot of targeting sick cells (a strategy, he says, that "could have been voodoo"). He also drank "an incredible" amount of fluids to help flush the toxins out of his system.
Side effects of the chemo, besides hair loss, included intense nausea, diarrhea, a loss of appetite, some severe lung toxicities that eventually resolved, and incredible fatigue.
"I've always been a runner, and I would even try to run while I was doing chemo," he said. "After I finished treatment, I would go literally 150 yards and just have to stop, and it took a lot of effort to work through it."
The experience reinforced his desire to stay in immunology, especially after having taken the most toxic chemotherapies.
"They worked, and I could tolerate them because I was young, but people who are older can't," Schreiber said. "The whole field of immunotherapy has really demonstrated that there are effective therapies out there that don't come with all of the same toxicities as the original chemo, so it was galvanizing to imagine contributing to finding some of those."
Schreiber went on to complete his MD and PhD degrees from the Sheila and David Fuente Program in Cancer Biology at the Miller School of Medicine and was nominated in 2011 as a Future Leader in Cancer Research by the American Association for Cancer Research. He also has numerous publications in the fields of tumor immunology and immunotherapy.
Sackstein, who was struck by Schreiber's enthusiasm and "boundless energy," predicts he will be a "major player in the world of therapeutics."
"The future for Taylor is amazing because he has the capacity to synthesize current knowledge and understand the gaps and then ask the right questions to establish new paradigms," said Sackstein, currently dean of the Herbert Wertheim College of Medicine at Florida International University. "It's a very unusual talent."
Since then, he has devoted his career to developing innovative techniques aimed at unleashing the immune system to attack cancer with less toxicity than chemotherapy and better clinical results—first, at a company called Heat Biologics and then at Pelican Therapeutics.
His primary work at Austin, Texas-based Shattuck is aimed at combining two functions in a single therapy for cancer and inflammatory diseases, blocking molecules that put a brake on the immune system (checkpoint inhibitors) while also stimulating the immune system's cancer-killing T cells.
The company has one drug in clinical testing as part of its Agonist Redirected Checkpoint (ARC) platform, which represents a new class of biological medicine. Two others are expected within the next year, with a pipeline of more than 250 drug candidates spanning cancer, inflammatory, and metabolic diseases.
Nine years after his own cancer diagnosis, Schreiber says it remains a huge part of his life, though his chances of a cancer recurrence today are about the same as his chances of getting newly diagnosed with any other cancer.
"I feel blessed to be in a position to help cancer patients live longer and could not imagine a more fulfilling way to spend my life," he says.

