New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
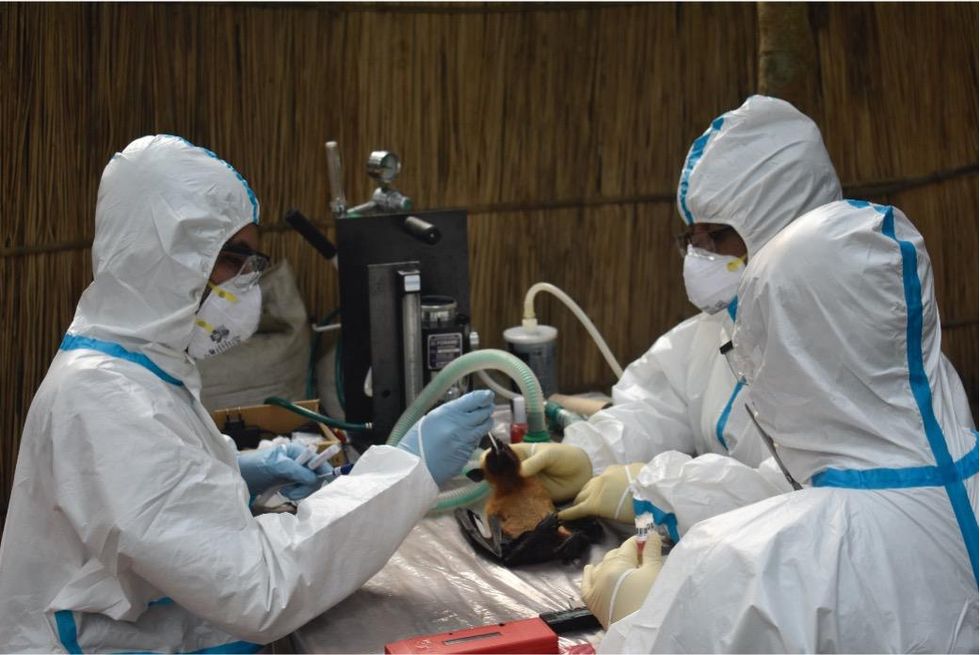
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
When Are We Obligated To Edit Wild Creatures?
Cows on a pasture, who, among other mammals, could experience immense suffering from the New World screwworm.
Combining CRISPR genome editing with the natural phenomenon of gene drive allows us to rewrite the genomes of wild organisms. The benefits of saving children from malaria by editing mosquitoes are obvious and much discussed, but humans aren't the only creatures who suffer. If we gain the power to intervene in a natural world "red in tooth and claw," yet decline to use it, are we morally responsible for the animal suffering that we could have prevented?
Given the power to alter the workings of the natural world, are we morally obligated to use it?
The scenario that may redefine our relationship with the natural world begins with fine clothing. You're dressed to the nines for a formal event, but you arrived early, and it's such a beautiful day that you decided to take a stroll by the nearby lake. Suddenly, you hear the sound of splashing and screams. A child is drowning! Will you dive in to save them? Or let them die, and preserve your expensive outfit?
The philosopher Peter Singer posited this scenario to show that we are all terrible human beings. Just about everyone would save the child and ruin the outfit... leading Singer to question why so few of us give equivalent amounts of money to save children on the other side of the world. The Against Malaria Foundation averages one life saved for every $7000.
But despite having a local bias, our moral compasses aren't completely broken. You never even considered letting the child drown because the situation wasn't your fault. That's because the cause of the problem simply isn't relevant: as the one who could intervene, the consequences are on your head. We are morally responsible for intervening in situations we did not create.
There is a critical difference between Singer's original scenario and the one above: in his version, it was a muddy pond. Any adult can rescue a child from a muddy pond, but a lake is different; you can only save the child if you know how to swim. We only become morally responsible when we acquire the power to intervene.
Few would disagree with either of these moral statements, but when they are combined with increasingly powerful technologies, the implications are deeply unsettling. Given the power to alter the workings of the natural world, are we morally obligated to use it? Recent developments suggest we had best determine the answer soon because, technologically, we are learning to swim. What choices will we make?
Gene drive is a natural phenomenon that occurs when a genetic element reliably spreads through a population even though it reduces the reproductive fitness of individual organisms. Nature has evolved many different mechanisms that result in gene drive, so many that it's nearly impossible to find an organism that doesn't have at least one driving element somewhere in its genome. More than half of our own DNA comprises the broken remnants of gene drives, plus a few active copies.
Scientists have long dreamed of harnessing gene drive to block mosquito-borne disease, with little success. Then came CRISPR genome editing, which works by cutting target genes and replacing them with a new sequence. What happens if you replace the original sequence with the edited version and an encoded copy of the CRISPR system? Gene drive.

CRISPR is a molecular scalpel that we can use to cut, and therefore replace, just about any DNA sequence in any cell. Encode the instructions for the CRISPR system adjacent to the new sequence, and genome editing will occur in the reproductive cells of subsequent generations of heterozygotes, always converting the original wild-type version to the new edited version. By ensuring that offspring will all be born of one sex, or by arranging for organisms that inherit two copies of the gene drive to be sterile, it's theoretically possible to cause a population crash.
(Credit: Esvelt)
When my colleagues and I first described this technology in 2014, we initially focused on the imperative for early transparency. Gene drive research is more like civic governance than traditional technology development: you can decline a treatment recommended by your doctor, but you can’t opt out when people change the shared environment. Applying the traditional closeted model of science to gene drive actively denies people a voice in decisions intended to affect them - and reforming scientific incentives for gene drive could be the first step to making all of science faster and safer.
But open gene drive research is clearly aligned with virtually all of our values. It's when technology places our deepest moral beliefs in conflict that we struggle, and learn who we truly are.
Two of our strongest moral beliefs include our reverence for the natural world and our abhorrence of suffering. Yet some natural species inherently cause tremendous suffering. Are we morally obligated to alter or even eradicate them?
To anyone who doubts that the natural world can inflict unimaginable suffering, consider the New World screwworm.
Judging by history, the answer depends on who is doing the suffering. We view the eradication of smallpox as one of our greatest triumphs, clearly demonstrating that we value human lives over the existence of disease-causing microorganisms. The same principle holds today for malaria: few would argue against using gene drive to crash populations of malarial mosquitoes to help eradicate the disease. There are more than 3500 species of mosquitoes, only three of which would be affected, and once malaria is gone, the mosquitoes could be allowed to recover. It would be extremely surprising if African nations decided not to eradicate malaria.
The more interesting question concerns our moral obligations to animals in the state of nature.
To anyone who doubts that the natural world can inflict unimaginable suffering, consider the New World screwworm, Cochyliomyia hominivorax. Female screwworm flies lay their eggs in open wounds, generating maggots that devour healthy tissue, gluttonously burrowing into the flesh of their host until they drop, engorged and sated, to metamorphose. Yet before they fall, the maggots in a wound emit a pheromone attracting new females, thereby acting as both conductors and performers in a macabre parade that consumes the host alive. The pain is utterly excruciating, so much so that infested people often require morphine before doctors can even examine the wound. Worst of all, the New World screwworm specializes in devouring complex mammals.
Every second of every day, hundreds of millions of animals suffer the excruciating agony of being eaten alive. It has been so throughout North and South America for millions of years. Until 2001, when humanity eradicated the last screwworm fly north of Panama using the “sterile insect techniqueâ€. This was not done to protect wild animals or even people, but for economic reasons: the cost of the program was small relative to the immense damage wrought by the screwworm on North American cattle, sheep, and goats. There were no obvious ecological effects. Despite being almost completely unknown even among animal rights activists, the screwworm elimination campaign may well have been one of the greatest triumphs of animal well-being.
Unfortunately, sterile insect technique isn't powerful enough to eradicate the screwworm from South America, where it is more entrenched and protected by the rougher terrain. But gene drive is.

Contrary to news hype, gene drive alone can't cause extinction, but if combined with conventional measures it might be possible to remove targeted species from the wild. For certain species that cause immense suffering, we may be morally obligated to do just that.
(Credit: Esvelt)
South Americans may well decide to eradicate screwworm for the same economic reasons that it was eradicated from North America: the fly inflicts $4 billion in annual damages on struggling rural communities that can least afford it. It need not go extinct, of course; the existence of the sterile insect facility in Panama proves that we can maintain the screwworm indefinitely in captivity on already dead meat.
Yet if for some reason humanity chooses to leave the screwworm as it is - even for upstanding moral reasons, whatever those may be - the knowledge of our responsibility should haunt us.
Tennyson wrote,
Are God and Nature then at strife,
That Nature lends such evil dreams?
So careful of the type she seems,
So careless of the single life.
Evolution by natural selection cares nothing for the single life, nor suffering, nor euphoria, save for their utility in replication. Theoretically, we do. But how much?
[Editor's Note: This story was originally published in May 2018. We are resurfacing archive hits while our staff is on vacation.]
A Fierce Mother vs. a Fatal Mutation
Amber Salzman, whose determination to find a cure for her son's rare disease led to a recently successful clinical trial using gene therapy.
Editor's Note: In the year 2000, Amber Salzman was a 39-year-old mom from Philadelphia living a normal life: working as a pharmaceutical executive, raising an infant son, and enjoying time with her family. But when tragedy struck in the form of a ticking time bomb in her son's DNA, she sprang into action. Her staggering triumphs after years of turmoil exemplify how parents today can play a crucial role in pushing science forward. This is her family's story, as told to LeapsMag's Editor-in-Chief Kira Peikoff.
For a few years, my nephew Oliver, suffered from symptoms that first appeared as attention deficit disorder and then progressed to what seemed like Asperger's, and he continued to worsen and lose abilities he once had. After repeated misdiagnoses, he was finally diagnosed at age 8 with adrenoleukodystrophy, or ALD – a degenerative brain disease that puts kids on the path toward death. We learned it was an X-linked disease, so we had to test other family members. Because Oliver had it, that meant his mother, my sister, was carrier, which meant I had a 50-50 chance of being a carrier, and if I was, then my son had a 50-50 chance of getting the bad gene.
You know how some people win prizes all the time? I don't have that kind of luck. I had a sick feeling when we drew my son's blood. It was almost late December in the year 2000. Spencer was 1 and climbing around like a monkey, starting to talk—a very rambunctious kid. He tested positive, along with Oliver's younger brother, Elliott.
"The only treatment at the time was an allogenic stem cell transplant from cord blood or bone marrow."
You can imagine the dreadful things that go through your mind. Everything was fine then, but he had a horrific chance that in about 3 or 4 years, a bomb would go off. It was so tough thinking that we were going to lose Oliver, and then Spencer and Elliott were next in line. The only treatment at the time was an allogenic stem cell transplant from cord blood or bone marrow, which required finding a perfect match in a donor and then undergoing months of excruciating treatment. The mortality rate can be as high as 40 percent. If your kid was lucky enough to find a donor, he would then be lucky to leave the hospital 100 days after a transplant with a highly fragile immune system.
At the time, I was at GlaxoSmithKline in Research and Development, so I did have a background in working with drug development and I was fortunate to report to the chairman of R&D, Tachi Yamada.
I called Tachi and said, "I need your advice, I have three or four years to find a cure. What do I do?" He did some research and said it's a monogenic disease—meaning it's caused by only one errant gene—so my best bet was gene therapy. This is an approach to treatment that involves taking a sample of the patient's own stem cells, treating them outside the body with a viral vector as a kind of Trojan Horse to deliver the corrected gene, and then infusing the solution back into the patient, in the hopes that the good gene will proliferate throughout the body and stop the disease in its tracks.
Tachi said to call his friend Jim Wilson, who was a leader in the field at UPenn.
Since I live in Philadelphia I drove to see Jim as soon as possible. What I didn't realize was how difficult a time it was. This was shortly after Jesse Gelsinger died in a clinical trial for gene therapy run by UPenn—the first death for the field—and research had abruptly stopped. But when I met with Jim, he provided a road map for what it would take to put together a gene therapy trial for ALD.
Meanwhile, in parallel, I was dealing with my son's health.
After he was diagnosed, we arranged a brain MRI to see if he had any early lesions, because the only way you can stop the disease is if you provide a bone marrow transplant before the disease evolves. Once it is in full force, you can't reverse it, like a locomotive that's gone wild.
"He didn't recover like other kids because his brain was not a normal brain; it was an ALD brain."
We found he had a brain tumor that had nothing to do with ALD. It was slow growing, and we would have never found it otherwise until it was much bigger and caused symptoms. Long story short, he ended up getting the tumor removed, and when he was healing, he didn't recover like other kids because his brain was not a normal brain; it was an ALD brain. We knew we needed a transplant soon, and the gene therapy trial was unfortunately still years away.
At the time, he was my only child, and I was thinking of having additional kids. But I didn't want to get pregnant with another ALD kid and I wanted a kid who could provide a bone marrow transplant for my son. So while my son was still OK, I went through 5 cycles of in vitro fertilization, a process in which hormone shots stimulated my ovaries to produce multiple eggs, which were then surgically extracted and fertilized in a lab with my husband's sperm. After the embryos grew in a dish for three to five days, doctors used a technique called preimplantation genetic diagnosis, screening those embryos to determine which genes they carry, in order to try to find a match for Spencer. Any embryo that had ALD, we saved for research. Any that did not have ALD but were not a match for Spencer, we put in the freezer. We didn't end up with a single one that was a match.
So he had a transplant at Duke Children's Hospital at age 2, using cord blood donated from a public bank. He had to be in the hospital a long time, infusing meds multiple times a day to prevent the donor cells from rejecting his body. We were all excited when he made it out after 100 days, but then we quickly had to go back for an infection he caught.
We were still bent on moving forward with the gene therapy trials.
Jim Wilson at Penn explained what proof of concept we needed in animals to go forward to humans, and a neurologist in Paris, Patrick Aubourg, had already done that using a vector to treat ALD mice. But he wasn't sure which vector to use in humans.
The next step was to get Patrick and a team of gene therapy experts together to talk about what they knew, and what needed to be done to get a trial started. There was a lot of talk about viral vectors. Because viruses efficiently transport their own genomes into the cells they infect, they can be useful tools for sending good genes into faulty cells. With some sophisticated tinkering, molecular biologists can neuter normally dangerous viruses to make them into delivery trucks, nothing more. The biggest challenge we faced then was: How do we get a viral vector that would be safe in humans?
Jim introduced us to Inder Verma, chair of the scientific advisory board of Cell Genesys, a gene therapy company in California that was focused on oncology. They were the closest to making a viral vector that could go into humans, based on a disabled form of HIV. When I spoke to Inder, he said, "Let's review the data, but you will need to convince the company to give you the vector." So I called the CEO and basically asked him, "Would you be willing to use the vector in this horrific disease?" I told him that our trial would be the fastest way to test their vector in humans. He said, "If you can convince my scientists this is ready to go, we will put the vector forward." Mind you, this was a multi-million-dollar commitment, pro bono.
I kept thinking every day, the clock is ticking, we've got to move quickly. But we convinced the scientists and got the vector.
Then, before we could test it, an unrelated clinical trial in gene therapy for a severe immunodeficiency disease, led to several of the kids developing leukemia in 2003. The press did a bad number and scared everyone away from the field, and the FDA put studies on hold in the U.S. That was one of those moments where I thought it was over. But we couldn't let it stop. Nothing's an obstacle, just a little bump we have to overcome.
Patrick wanted to do the study in France with the vector. This is where patient advocacy is important in providing perspective on the risks vs. benefits of undergoing an experimental treatment. What nobody seemed to realize was that the kids in the 2003 trial would have died if they were not first given the gene therapy, and luckily their leukemia was a treatable side effect.
Patrick and I refused to give up pushing for approval of the trial in France. Meanwhile, I was still at GSK, working full time, and doing this at night, nonstop. Because my day job did require travel to Europe, I would stop by Paris and meet with him. Another sister of mine who did not have any affected children was a key help and we kept everything going. You really need to continually stay engaged and press the agenda forward, since there are so many things that pop up that can derail the program.
Finally, Patrick was able to treat four boys with the donated vector. The science paper came out in 2009. It was a big deal. That's when the venture money came in—Third Rock Ventures was the first firm to put big money behind gene therapy. They did a deal with Patrick to get access to the Intellectual Property to advance the trial, brought on scientists to continue the study, and made some improvements to the vector. That's what led to the new study reported recently in the New England Journal of Medicine. Of 17 patients, 15 of them are still fine at least two years after treatment.
You know how I said we felt thrilled that my son could leave the hospital after 100 days? When doing the gene therapy treatment, the hospital stay needed is much quicker. Shortly after one kid was treated, a physician in the hospital remarked, "He is fine, he's only here because of the trial." Since you get your own cells, there is no risk of graft vs. host disease. The treatment is pretty anticlimactic: a bag of blood, intravenously infused. You can bounce back within a few weeks.
Now, a few years out, approximately 20 percent of patients' cells have been corrected—and that's enough to hold off the disease. That's what the data is showing. I was blown away when it worked in the first two patients.
The formerly struggling field is now making a dramatic comeback.
Just last month, the first two treatments involving gene therapy were approved by the FDA to treat a devastating type of leukemia in children and an aggressive blood cancer in adults.
Now I run a company, Adverum Biotechnologies, that I wish existed back when my son was diagnosed, because I want people who are like me, coming to me, saying: "I have proof of concept in an animal, I need to get a vector suitable for human trials, do the work needed to file with the FDA, and move it into humans." Our company knows how to do that and would like to work with such patient advocates.
Often parents feel daunted to partake in similar efforts, telling me, "Well, you worked in pharma." Yes, I had advantages, but if you don't take no for an answer, people will help you. Everybody is one degree of separation from people who can help them. You don't need a science or business background. Just be motivated, ask for help, and have your heart in the right place.
Having said that, I don't want to sound judgmental of families who are completely paralyzed. When you get a diagnosis that your child is dying, it is hard to get out of bed in the morning and face life. My sister at a certain point had one child dying, one in the hospital getting a transplant, and a healthy younger child. To expect someone like that to at the same time be flying to an FDA meeting, it's hard. Yet, she made critical meetings, and she and her husband graciously made themselves available to talk to parents of recently diagnosed boys. But it is really tough and my heart goes out to anyone who has to live through such devastation.
Tragically, my nephew Oliver passed away 13 years ago at age 12. My other nephew was 8 when he had a cord blood transplant; our trial wasn't available yet. He had some bad graft vs. host disease and he is now navigating life using a wheelchair, but thank goodness, it stopped the disease. He graduated Stanford a year ago and is now a sports writer for the Houston Chronicle.
As for my son, today he is 17, a precocious teenager applying to colleges. He also volunteers for an organization called the Friendship Circle, providing friends for kids with special needs. He doesn't focus on disability and accepts people for who they are – maybe he would have been like that anyway, but it's part of who he is. He lost his cousin and knows he is alive today because Oliver's diagnosis gave us a head start on his.
My son's story is a good one in that he had a successful transplant and recovered.
Once we knew he would make it and we no longer needed our next child to be a match, we had a daughter using one of our healthy IVF embryos in storage. She is 14 now, but she jokes that she is technically 17, so she should get to drive. I tell her, they don't count the years in the freezer. You have to joke about it.
I am so lucky to have two healthy kids today based on advances in science.
And I often think of Oliver. We always try to make him proud and honor his name.
[Editor's Note: This story was originally published in November 2017. We are resurfacing archive hits while our staff is on vacation.]

Salzman and her son Spencer, 17, who is now healthy.
(Courtesy of Salzman)

