New tools could catch disease outbreaks earlier - or predict them

A team at the Catalan Institution for Research and Advanced Studies is utilizing drones and weather stations to collect data on how mosquito breeding patterns are changing in response to climate shifts.
Every year, the villages which lie in the so-called ‘Nipah belt’— which stretches along the western border between Bangladesh and India, brace themselves for the latest outbreak. For since 1998, when Nipah virus—a form of hemorrhagic fever most common in Bangladesh—first spilled over into humans, it has been a grim annual visitor to the people of this region.
With a 70 percent fatality rate, no vaccine, and no known treatments, Nipah virus has been dubbed in the Western world as ‘the worst disease no one has ever heard of.’ Currently, outbreaks tend to be relatively contained because it is not very transmissible. The virus circulates throughout Asia in fruit eating bats, and only tends to be passed on to people who consume contaminated date palm sap, a sweet drink which is harvested across Bangladesh.
But as SARS-CoV-2 has shown the world, this can quickly change.
“Nipah virus is among what virologists call ‘the Big 10,’ along with things like Lassa fever and Crimean Congo hemorrhagic fever,” says Noam Ross, a disease ecologist at New York-based non-profit EcoHealth Alliance. “These are pretty dangerous viruses from a lethality perspective, which don’t currently have the capacity to spread into broader human populations. But that can evolve, and you could very well see a variant emerge that has human-human transmission capability.”
That’s not an overstatement. Surveys suggest that mammals harbour about 40,000 viruses, with roughly a quarter capable of infecting humans. The vast majority never get a chance to do so because we don’t encounter them, but climate change can alter that. Recent studies have found that as animals relocate to new habitats due to shifting environmental conditions, the coming decades will bring around 300,000 first encounters between species which normally don’t interact, especially in tropical Africa and southeast Asia. All these interactions will make it far more likely for hitherto unknown viruses to cross paths with humans.
That’s why for the last 16 years, EcoHealth Alliance has been conducting ongoing viral surveillance projects across Bangladesh. The goal is to understand why Nipah is so much more prevalent in the western part of the country, compared to the east, and keep a watchful eye out for new Nipah strains as well as other dangerous pathogens like Ebola.
"There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them," says Cat Lippi, medical geography researcher at the University of Florida.
Until very recently this kind of work has been hampered by the limitations of viral surveillance technology. The PREDICT project, a $200 million initiative funded by the United States Agency for International Development, which conducted surveillance across the Amazon Basin, Congo Basin and extensive parts of South and Southeast Asia, relied upon so-called nucleic acid assays which enabled scientists to search for the genetic material of viruses in animal samples.
However, the project came under criticism for being highly inefficient. “That approach requires a big sampling effort, because of the rarity of individual infections,” says Ross. “Any particular animal may be infected for a couple of weeks, maybe once or twice in its lifetime. So if you sample thousands and thousands of animals, you'll eventually get one that has an Ebola virus infection right now.”
Ross explains that there is now far more interest in serological sampling—the scientific term for the process of drawing blood for antibody testing. By searching for the presence of antibodies in the blood of humans and animals, scientists have a greater chance of detecting viruses which started circulating recently.
Despite the controversy surrounding EcoHealth Alliance’s involvement in so-called gain of function research—experiments that study whether viruses might mutate into deadlier strains—the organization’s separate efforts to stay one step ahead of pathogen evolution are key to stopping the next pandemic.
“Having really cheap and fast surveillance is really important,” says Ross. “Particularly in a place where there's persistent, low level, moderate infections that potentially have the ability to develop into more epidemic or pandemic situations. It means there’s a pathway that something more dangerous can come through."
Scientists are searching for the presence of antibodies in the blood of humans and animals in hopes to detect viruses that recently started circulating.
EcoHealth Alliance
In Bangladesh, EcoHealth Alliance is attempting to do this using a newer serological technology known as a multiplex Luminex assay, which tests samples against a panel of known antibodies against many different viruses. It collects what Ross describes as a ‘footprint of information,’ which allows scientists to tell whether the sample contains the presence of a known pathogen or something completely different and needs to be investigated further.
By using this technology to sample human and animal populations across the country, they hope to gain an idea of whether there are any novel Nipah virus variants or strains from the same family, as well as other deadly viral families like Ebola.
This is just one of several novel tools being used for viral discovery in surveillance projects around the globe. Multiple research groups are taking PREDICT’s approach of looking for novel viruses in animals in various hotspots. They collect environmental DNA—mucus, faeces or shed skin left behind in soil, sediment or water—which can then be genetically sequenced.
Five years ago, this would have been a painstaking work requiring bringing collected samples back to labs. Today, thanks to the vast amounts of money spent on new technologies during COVID-19, researchers now have portable sequencing tools they can take out into the field.
Christopher Jerde, a researcher at the UC Santa Barbara Marine Science Institute, points to the Oxford Nanopore MinION sequencer as one example. “I tried one of the early versions of it four years ago, and it was miserable,” he says. “But they’ve really improved, and what we’re going to be able to do in the next five to ten years will be amazing. Instead of having to carefully transport samples back to the lab, we're going to have cigar box-shaped sequencers that we take into the field, plug into a laptop, and do the whole sequencing of an organism.”
In the past, viral surveillance has had to be very targeted and focused on known families of viruses, potentially missing new, previously unknown zoonotic pathogens. Jerde says that the rise of portable sequencers will lead to what he describes as “true surveillance.”
“Before, this was just too complex,” he says. “It had to be very focused, for example, looking for SARS-type viruses. Now we’re able to say, ‘Tell us all the viruses that are here?’ And this will give us true surveillance – we’ll be able to see the diversity of all the pathogens which are in these spots and have an understanding of which ones are coming into the population and causing damage.”
But being able to discover more viruses also comes with certain challenges. Some scientists fear that the speed of viral discovery will soon outpace the human capacity to analyze them all and assess the threat that they pose to us.
“I think we're already there,” says Jason Ladner, assistant professor at Northern Arizona University’s Pathogen and Microbiome Institute. “If you look at all the papers on the expanding RNA virus sphere, there are all of these deposited partial or complete viral sequences in groups that we just don't know anything really about yet.” Bats, for example, carry a myriad of viruses, whose ability to infect human cells we understand very poorly.
Cultivating these viruses under laboratory conditions and testing them on organoids— miniature, simplified versions of organs created from stem cells—can help with these assessments, but it is a slow and painstaking work. One hope is that in the future, machine learning could help automate this process. The new SpillOver Viral Risk Ranking platform aims to assess the risk level of a given virus based on 31 different metrics, while other computer models have tried to do the same based on the similarity of a virus’s genomic sequence to known zoonotic threats.
However, Ladner says that these types of comparisons are still overly simplistic. For one thing, scientists are still only aware of a few hundred zoonotic viruses, which is a very limited data sample for accurately assessing a novel pathogen. Instead, he says that there is a need for virologists to develop models which can determine viral compatibility with human cells, based on genomic data.
“One thing which is really useful, but can be challenging to do, is understand the cell surface receptors that a given virus might use,” he says. “Understanding whether a virus is likely to be able to use proteins on the surface of human cells to gain entry can be very informative.”
As the Earth’s climate heats up, scientists also need to better model the so-called vector borne diseases such as dengue, Zika, chikungunya and yellow fever. Transmitted by the Aedes mosquito residing in humid climates, these blights currently disproportionally affect people in low-income nations. But predictions suggest that as the planet warms and the pests find new homes, an estimated one billion people who currently don’t encounter them might be threatened by their bites by 2080. “When it comes to mosquito-borne diseases we have to worry about shifts in suitable habitat,” says Cat Lippi, a medical geography researcher at the University of Florida. “As climate patterns change on these big scales, we expect to see shifts in where people will be at risk for contracting these diseases.”
Public health practitioners and government decision-makers need tools to make climate-informed decisions about the evolving threat of different infectious diseases. Some projects are already underway. An ongoing collaboration between the Catalan Institution for Research and Advanced Studies and researchers in Brazil and Peru is utilizing drones and weather stations to collect data on how mosquitoes change their breeding patterns in response to climate shifts. This information will then be fed into computer algorithms to predict the impact of mosquito-borne illnesses on different regions.

The team at the Catalan Institution for Research and Advanced Studies is using drones and weather stations to collect data on how mosquito breeding patterns change due to climate shifts.
Gabriel Carrasco
Lippi says that similar models are urgently needed to predict how changing climate patterns affect respiratory, foodborne, waterborne and soilborne illnesses. The UK-based Wellcome Trust has allocated significant assets to fund such projects, which should allow scientists to monitor the impact of climate on a much broader range of infections. “There are a lot of different infectious agents that are sensitive to climate change that don't have these sorts of software tools being developed for them,” she says.
COVID-19’s havoc boosted funding for infectious disease research, but as its threats begin to fade from policymakers’ focus, the money may dry up. Meanwhile, scientists warn that another major infectious disease outbreak is inevitable, potentially within the next decade, so combing the planet for pathogens is vital. “Surveillance is ultimately a really boring thing that a lot of people don't want to put money into, until we have a wide scale pandemic,” Jerde says, but that vigilance is key to thwarting the next deadly horror. “It takes a lot of patience and perseverance to keep looking.”
This article originally appeared in One Health/One Planet, a single-issue magazine that explores how climate change and other environmental shifts are increasing vulnerabilities to infectious diseases by land and by sea. The magazine probes how scientists are making progress with leaders in other fields toward solutions that embrace diverse perspectives and the interconnectedness of all lifeforms and the planet.
Can You Trust Your Gut for Food Advice?
Which foods are actually healthy for your individual gut microbiome? Several companies are offering personalized dietary guidance based on your test results, but their answers in one experiment turned up with some conflicting advice.
I recently got on the scale to weigh myself, thinking I've got to eat better. With so many trendy diets today claiming to improve health, from Keto to Paleo to Whole30, it can be confusing to figure out what we should and shouldn't eat for optimal nutrition.
A number of companies are now selling the concept of "personalized" nutrition based on the genetic makeup of your individual gut bugs.
My next thought was: I've got to lose a few pounds.
Consider a weird factoid: In addition to my fat, skin, bone and muscle, I'm carrying around two or three pounds of straight-up bacteria. Like you, I am the host to trillions of micro-organisms that live in my gut and are collectively known as my microbiome. An explosion of research has occurred in the last decade to try to understand exactly how these microbial populations, which are unique to each of us, may influence our overall health and potentially even our brains and behavior.
Lots of mysteries still remain, but it is established that these "bugs" are crucial to keeping our body running smoothly, performing functions like stimulating the immune system, synthesizing important vitamins, and aiding digestion. The field of microbiome science is evolving rapidly, and a number of companies are now selling the concept of "personalized" nutrition based on the genetic makeup of your individual gut bugs. The two leading players are Viome and DayTwo, but the landscape includes the newly launched startup Onegevity Health and others like Thryve, which offers customized probiotic supplements in addition to dietary recommendations.
The idea has immediate appeal – if science could tell you exactly what to make for lunch and what to avoid, you could forget about the fad diets and go with your own bespoke food pyramid. Wondering if the promise might be too good to be true, I decided to perform my own experiment.
Last fall, I sent the identical fecal sample to both Viome (I paid $425, but the price has since dropped to $299) and DayTwo ($349). A couple of months later, both reports finally arrived, and I eagerly opened each app to compare their recommendations.
First, I examined my results from Viome, which was founded in 2016 in Cupertino, Calif., and declares without irony on its website that "conflicting food advice is now obsolete."
I learned I have "average" metabolic fitness and "average" inflammatory activity in my gut, which are scores that the company defines based on a proprietary algorithm. But I have "low" microbial richness, with only 62 active species of bacteria identified in my sample, compared with the mean of 157 in their test population. I also received a list of the specific species in my gut, with names like Lactococcus and Romboutsia.
But none of it meant anything to me without actionable food advice, so I clicked through to the Recommendations page and found a list of My Superfoods (cranberry, garlic, kale, salmon, turmeric, watermelon, and bone broth) and My Foods to Avoid (chickpeas, kombucha, lentils, and rice noodles). There was also a searchable database of many foods that had been categorized for me, like "bell pepper; minimize" and "beef; enjoy."
"I just don't think sufficient data is yet available to make reliable personalized dietary recommendations based on one's microbiome."
Next, I looked at my results from DayTwo, which was founded in 2015 from research out of the Weizmann Institute of Science in Israel, and whose pitch to consumers is, "Blood sugar made easy. The algorithm diet personalized to you."
This app had some notable differences. There was no result about my metabolic fitness, microbial richness, or list of the species in my sample. There was also no list of superfoods or foods to avoid. Instead, the app encouraged me to build a meal by searching for foods in their database and combining them in beneficial ways for my blood sugar. Two slices of whole wheat bread received a score of 2.7 out of 10 ("Avoid"), but if combined with one cup of large curd cottage cheese, the score improved to 6.8 ("Limit"), and if I added two hard-boiled eggs, the score went up to 7.5 ("Good").
Perusing my list of foods with "Excellent" scores, I noticed some troubling conflicts with the other app. Lentils, which had been a no-no according to Viome, received high marks from DayTwo. Ditto for Kombucha. My purported superfood of cranberry received low marks. Almonds got an almost perfect score (9.7) while Viome told me to minimize them. I found similarly contradictory advice for foods I regularly eat, including navel oranges, peanuts, pork, and beets.
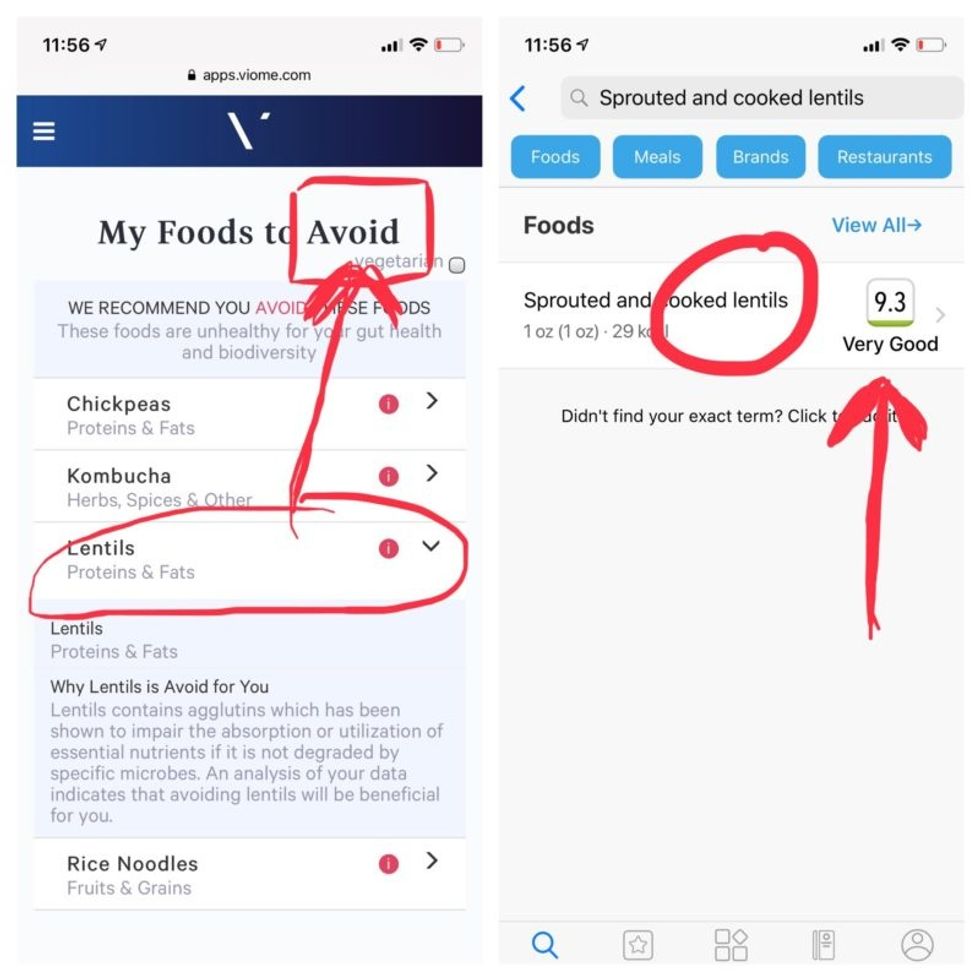
Contradictory dietary guidance that Kira Peikoff received from Viome (left) and DayTwo from an identical sample.
To be sure, there was some overlap. Both apps agreed on rice noodles (bad), chickpeas (bad), honey (bad), carrots (good), and avocado (good), among other foods.
But still, I was left scratching my head. Which set of recommendations should I trust, if either? And what did my results mean for the accuracy of this nascent field?
I called a couple of experts to find out.
"I have worked on the microbiome and nutrition for the last 20 years and I would be absolutely incapable of finding you evidence in the scientific literature that lentils have a detrimental effect based on the microbiome," said Dr. Jens Walter, an Associate Professor and chair for Nutrition, Microbes, and Gastrointestinal Health at the University of Alberta. "I just don't think sufficient data is yet available to make reliable personalized dietary recommendations based on one's microbiome. And even if they would have proprietary algorithms, at least one of them is not doing it right."
There is definite potential for personalized nutrition based on the microbiome, he said, but first, predictive models must be built and standardized, then linked to clinical endpoints, and tested in a large sample of healthy volunteers in order to enable extrapolations for the general population.
"It is mindboggling what you would need to do to make this work," he observed. "There are probably hundreds of relevant dietary compounds, then the microbiome has at least a hundred relevant species with a hundred or more relevant genes each, then you'd have to put all this together with relevant clinical outcomes. And there's a hundred-fold variation in that information between individuals."
However, Walter did acknowledge that the companies might be basing their algorithms on proprietary data that could potentially connect all the dots. I reached out to them to find out.
Amir Golan, the Chief Commercial Officer of DayTwo, told me, "It's important to emphasize this is a prediction, as the microbiome field is in a very early stage of research." But he added, "I believe we are the only company that has very solid science published in top journals and we can bring very actionable evidence and benefit to our uses."
He was referring to pioneering work out of the Weizmann Institute that was published in 2015 in the journal Cell, which logged the glycemic responses of 800 people in response to nearly 50,000 meals; adding information about the subjects' microbiomes enabled more accurate glycemic response predictions. Since then, Golan said, additional trials have been conducted, most recently with the Mayo Clinic, to duplicate the results, and other studies are ongoing whose results have not yet been published.
He also pointed out that the microbiome was merely one component that goes into building a client's profile, in addition to medical records, including blood glucose levels. (I provided my HbA1c levels, a measure of average blood sugar over the previous several months.)
"We are not saying we want to improve your gut microbiome. We provide a dynamic tool to help guide what you should eat to control your blood sugar and think about combinations," he said. "If you eat one thing, or with another, it will affect you in a different way."
Viome acknowledged that the two companies are taking very different approaches.
"DayTwo is primarily focused on the glycemic response," Naveen Jain, the CEO, told me. "If you can only eat butter for rest of your life, you will have no glycemic response but will probably die of a heart attack." He laughed. "Whereas we came from very different angle – what is happening inside the gut at a microbial level? When you eat food like spinach, how will that be metabolized in the gut? Will it produce the nutrients you need or cause inflammation?"
He said his team studied 1000 people who were on continuous glucose monitoring and fed them 45,000 meals, then built a proprietary data prediction model, looking at which microbes existed and how they actively broke down the food.
Jain pointed out that DayTwo sequences the DNA of the microbes, while Viome sequences the RNA – the active expression of DNA. That difference, in his opinion, is key to making accurate predictions.
"DNA is extremely stable, so when you eat any food and measure the DNA [in a fecal sample], you get all these false positives--you get DNA from plant food and meat, and you have no idea if those organisms are dead and simply transient, or actually exist. With RNA, you see what is actually alive in the gut."
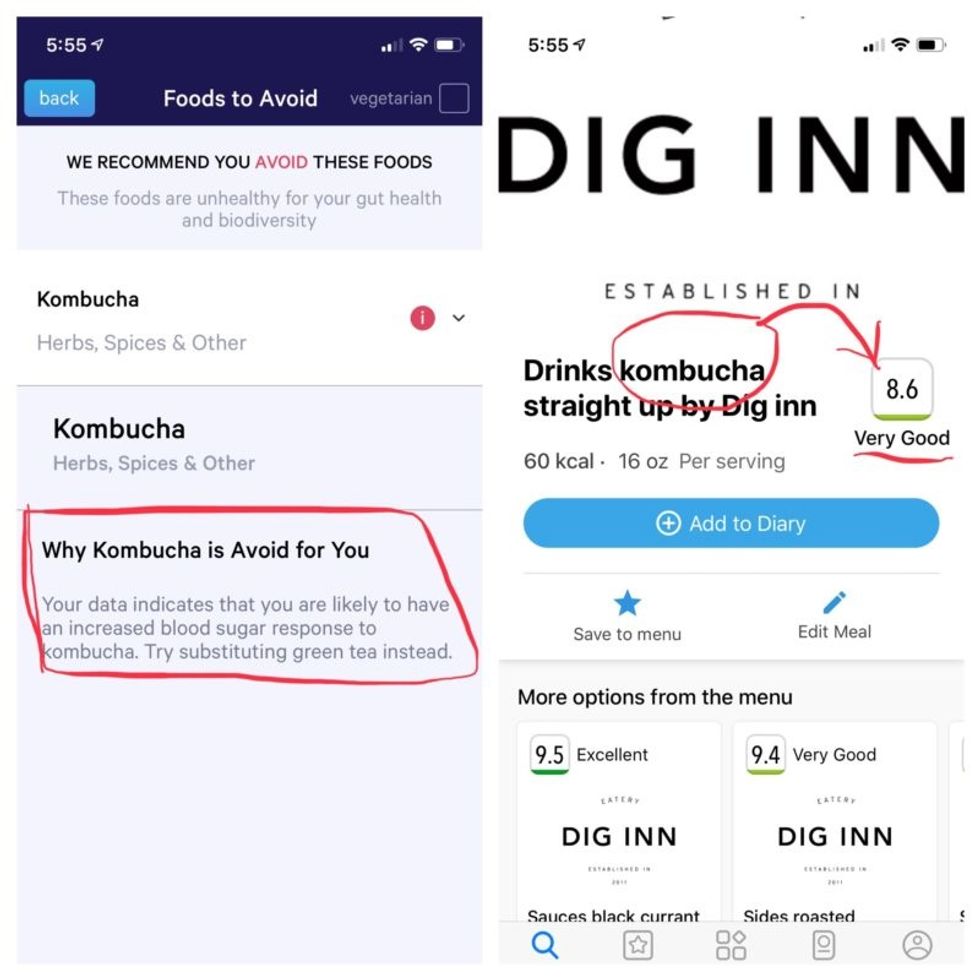
More contradictory food advice from Viome (left) and DayTwo.
Note that controversy exists over how it is possible with a fecal sample to effectively measure RNA, which degrades within minutes, though Jain said that his company has the technology to keep RNA stable for fourteen days.
Viome's approach, Jain maintains, is 90 percent accurate, based on as-yet unpublished data; a patent was filed just last week. DayTwo's approach is 66 percent accurate according to the latest published research.
Natasha Haskey, a registered dietician and doctoral student conducting research in the field of microbiome science and nutrition, is skeptical of both companies. "We can make broad statements, like eat more fruits and vegetables and fiber, but when it comes to specific foods, the science is just not there yet," she said. "I think there is a future, and we will be doing that someday, but not yet. Maybe we will be closer in ten years."
Professor Walter wholeheartedly agrees with Haskey, and suggested that if people want to eat a gut-healthy diet, they should focus on beneficial oils, fruits and vegetables, fish, a variety of whole grains, poultry and beans, and limit red meat and cheese, as well as avoid processed meats.
"These services are far over the tips of their science skis," Arthur Caplan, the founding head of New York University's Division of Medical Ethics, said in an email. "We simply don't know enough about the gut microbiome, its fluctuations and variability from person to person to support general [direct-to-consumer] testing. This is simply premature. We need standards for accuracy, specificity, and sensitivity, plus mandatory competent counseling for all such testing. They don't exist. Neither should DTC testing—yet."
Meanwhile, it's time for lunch. I close out my Viome and DayTwo apps and head to the kitchen to prepare a peanut butter sandwich. My gut tells me I'll be just fine.
Kira Peikoff was the editor-in-chief of Leaps.org from 2017 to 2021. As a journalist, her work has appeared in The New York Times, Newsweek, Nautilus, Popular Mechanics, The New York Academy of Sciences, and other outlets. She is also the author of four suspense novels that explore controversial issues arising from scientific innovation: Living Proof, No Time to Die, Die Again Tomorrow, and Mother Knows Best. Peikoff holds a B.A. in Journalism from New York University and an M.S. in Bioethics from Columbia University. She lives in New Jersey with her husband and two young sons. Follow her on Twitter @KiraPeikoff.
Should You Bank Your Kid’s Teeth for Stem Cells?
When Karen Davis attended a presentation at a dental conference in 2013, she unexpectedly discovered a service that could help her daughter, Madeline: storing stem cells derived from her teeth that potentially could be used in the future to treat her Crohn's disease.
"Even though this isn't a viable option today, I know how rapidly things can change."
Throughout high school, Madeline suffered from the painful autoimmune disorder, which wreaks havoc on the gastrointestinal system and can lead to life-threatening complications.
"I leave no stone unturned when it comes to medical care and this resonated with me," says Davis, a Dallas-based dental hygienist who was encouraged by advances in stem cell research. Later that year, when Madeline got her wisdom teeth extracted, Davis shipped them off to the Store-A-Tooth company in Massachusetts, where they will be kept frozen until needed. "Even though this isn't a viable option today, I know how rapidly things can change," says Davis. "To me, this was a worthwhile investment—I didn't want to miss out on an opportunity that would provide a pathway to a cure."

Karen Davis pictured with her daughter Madeline.
(Courtesy of Karen Davis)
The process itself was straightforward. Madeline's newly extracted wisdom teeth--baby teeth can be saved, too—were bathed in a special solution, loaded into a Styrofoam container lined with cold packs and sent to the stem cell company. There, a team harvested the dental stem cells from the pulp, then grew them in culture and cryogenically preserved them. Store-A-Tooth charges $1500-1749 for tooth collection and $120 per year for storage, while other dental pulp stem cell tissue banks cost $500-$600 upfront and in the $120 range annually for storage.
The rationale here is that if you missed out on banking your baby's umbilical cord blood, this gives you another chance to harvest their stem cells. "If their child later develops an illness that could be managed or even cured with stem cell therapy, this is an insurance policy," says Amr Moursi, DDS, PhD, chair of the department of pediatric dentistry at New York University College of Dentistry.
But is there a genuine potential here for some effective treatments in the relatively near future—or is this just another trendy fad? Scientific opinion is decidedly mixed. Stem cells have been heralded as the next frontier in medicine because of their versatility: with a little chemical coaxing, they can be transformed into different cell types, such as heart, blood or brain cells, to create tissue that can mend damaged body parts. Because they're taken from your own body, there's little chance of rejection, which means patients don't have to take strong antirejection drugs that can have all sorts of unpleasant side effects for the rest of their lives.
However, while stem cells are immature cells found in different tissues, ranging from abdominal fat to bone marrow, there is a vast difference between the stem cells found in cord blood and in teeth. Cord blood, which is culled from the umbilical cord when a baby is born, contains what are called hematopoietic stem cells (HSCs), which can mature into other blood cells. These type of stem cells have already been approved by the U.S. Food and Drug Administration to treat patients—especially children--with blood cancers, such as leukemias and lymphomas, and certain blood disorders like sickle cell anemia.
In contrast, stem cells in teeth are called mesenchymal stem cells (MSCs), which are found in dental pulp, the tissue in the center of the tooth that's filled with nerves and blood vessels. MSCs are adult stem cells normally found in the bone marrow that can transform into bone, fat, and cartilage cells, and also aid in the formation of blood stem cells.
"Right now we just don't have rigorous evidence that they can be used in that fashion and have real benefit."
Small studies on lab animals suggest that MSCs secrete growth factors—hormonal steroids or proteins—that can nurture ailing cells, act as powerful anti-inflammatory agents that could tame autoimmune disorders like the one that plagues Karen Davis's daughter, and may even generate new nerve and muscle tissue. Preliminary research suggests they potentially could treat medical conditions as varied as heart disease, spinal cord injury and type 1 diabetes by generating new cells, which can replace damaged or dead cells.
But this is all very early research and there's a vast difference between how cells behave in the tightly controlled environment of a lab versus the real world in a diverse population of human patients. "Right now we just don't have rigorous evidence that they can be used in that fashion and have real benefit," says Pamela G. Robey, PhD, chief of the skeletal biology section at the National Institute of Dental and Craniofacial Research at the National Institutes of Health.
Robey should know—she headed the research team that discovered stem cells in human baby teeth and in wisdom teeth more than fifteen years ago. She believes prospects are better using these stem cells for tooth repair: research suggests they may be able to fix cracked teeth, repair bone defects caused by gum disease, or in root canal therapy, where they can be used to replace infected tissue with regenerated healthy pulp.
In the meantime, though, there are no clinical applications for MSCs. "These tooth banking companies aren't doing their own research," says Leigh Turner, a bioethicist at the University of Minnesota who monitors stem cell clinics. "They cobble together reports of early research in humans or from animal studies in an effort to provide a narrative to make it seem like it is evidence based."
Still, in all fairness, tooth banking companies aren't making the kind of extravagant claims made by stem cell clinics, which operate in a gray area of the law and purport to treat everything from chronic lung disease to Alzheimer's. "We don't know when therapies will be available using these cells because the pace of research is hard to predict," says Peter Verlander, PhD, a molecular geneticist and chief scientific officer of Provia Laboratories, the parent company of Store-A-Tooth. "But for parents who regretted not banking their child's cord blood, especially if they later develop a disease like diabetes, this is another opportunity."
But the jury is still out if this is truly a good investment. Moursi, a national spokesperson for the American Academy of Pediatric Dentistry who fields queries about this practice from a dozen or so families a year, concludes: "If you could afford it, and know the risks, benefits and current limitations, then it is something to consider."

