The Dangers of Hype: How a Bold Claim and Sensational Media Unraveled a Company
Magnetic resonance imaging of the brain.
This past March, headlines suddenly flooded the Internet about a startup company called Nectome. Founded by two graduates of the Massachusetts Institute of Technology, the new company was charging people $10,000 to join a waiting list to have their brains embalmed, down to the last neuron, using an award-winning chemical compound.
While the lay public presumably burnt their wills and grew ever more excited about the end of humanity's quest for immortality, neurologists let out a collective sigh.
Essentially, participants' brains would turn to a substance like glass and remain in a state of near-perfect preservation indefinitely. "If memories can truly be preserved by a sufficiently good brain banking technique," Nectome's website explains, "we believe that within the century it could become feasible to digitize your preserved brain and use that information to recreate your mind." But as with most Faustian bargains, Nectome's proposition came with a serious caveat -- death.
That's right, in order for Nectome's process to properly preserve your connectome, the comprehensive map of the brain's neural connections, you must be alive (and under anesthesia) while the fluid is injected. This way, the company postulates, when the science advances enough to read and extract your memories someday, your vitrified brain will still contain your perfectly preserved essence--which can then be digitally recreated as a computer simulation.
Almost immediately this story gained buzz with punchy headlines: "Startup wants to upload your brain to the cloud, but has to kill you to do it," "San Junipero is real: Nectome wants to upload your brain," and "New tech firm promises eternal life, but you have to die."
While the lay public presumably burnt their wills and grew ever more excited about the end of humanity's quest for immortality, neurologists let out a collective sigh -- hype had struck the scientific community once again.
The truth about Nectome is that its claims are highly speculative and no hard science exists to suggest that our connectome is the key to our 'being,' nor that it can ever be digitally revived. "We haven't come even close to understanding even the most basic types of functioning in the brain," says neuroscientist Alex Fox, who was educated at the University of Queensland in Australia. "Memory storage in the brain is only a theoretical concept [and] there are some seriously huge gaps in our knowledge base that stand in the way of testing [the connectome] theory."
After the Nectome story broke, Harvard computational neuroscientist Sam Gershman tweeted out:
"Didn't anyone tell them that we've known the C Elegans (a microscopic worm) connectome for over a decade but haven't figured out how to reconstruct all of their memories? And that's only 7000 synapses compared to the trillions of synapses in the human brain!"
Hype can come from researchers themselves, who are under an enormous amount of pressure to publish original work and maintain funding.
How media coverage of Nectome went from an initial fastidiously researched article in the MIT Technology Review by veteran science journalist Antonio Regalado to the click-bait frenzy it became is a prime example of the 'science hype' phenomenon. According to Adam Auch, who holds a doctorate in philosophy from Dalhousie University in Nova Scotia, Canada, "Hype is a feature of all stages of the scientific dissemination process, from the initial circulation of preliminary findings within particular communities of scientists, to the process by which such findings come to be published in peer-reviewed journals, to the subsequent uptake these findings receive from the non-specialist press and the general public."
In the case of Nectome, hype was present from the word go. Riding the high of several major wins, including having raised over one million dollars in funding and partnering with well-known MIT neurologist Edward Boyden, Nectome founders Michael McCanna and Robert McIntyre launched their website on March 1, 2018. Just one month prior, they were able to purchase and preserve a newly deceased corpse in Portland, Oregon, showing that vitrifixation, their method of chemical preservation, could be used on a human specimen. It had previously won an award for preserving every synaptic structure on a rabbit brain.
The Nectome mission statement, found on its website, is laced with saccharine language that skirts the unproven nature of the procedure the company is peddling for big bucks: "Our mission is to preserve your brain well enough to keep all its memories intact: from that great chapter of your favorite book to the feeling of cold winter air, baking an apple pie, or having dinner with your friends and family."
This rhetoric is an example of hype that can come from researchers themselves, who are under an enormous amount of pressure to publish original work and maintain funding. As a result, there is a constant push to present science as "groundbreaking" when really, as is apparently the case with Nectome, it is only a small piece in a much larger effort.
Calling out the audacity of Nectome's posited future, neuroscientist Gershman commented to another publication, "The important question is whether the connectome is sufficient for memory: Can I reconstruct all memories knowing only the connections between neurons? The answer is almost certainly no, given our knowledge about how memories are stored (itself a controversial topic)."

The former home page of Nectome's website, which has now been replaced by a statement titled, "Response to recent press."
Furthermore, universities like MIT, who entered into a subcontract with Nectome, are under pressure to seek funding through partnerships with industry as a result of the Bayh-Dole Act of 1980. Also known as the Patent and Trademark Law Amendments Act, this piece of legislation allows universities to commercialize inventions developed under federally funded research programs, like Nectome's method of preserving brains, formally called Aldehyde-Stabilized Cryopreservation.
"[Universities use] every incentive now to talk about innovation," explains Dr. Ivan Oransky, president of the Association of Health Care Journalists and co-founder of retractionwatch.com, a blog that catalogues errors and fraud in published research. "Innovation to me is often a fancy word for hype. The role of journalists should not be to glorify what universities [say, but to] tell the closest version of the truth they can."
In this case, a combination of the hyperbolic press, combined with some impressively researched expose pieces, led MIT to cut its ties with Nectome on April 2nd, 2018, just two weeks after the news of their company broke.
The solution to the dangers of hype, experts say, is a more scientifically literate public—and less clickbait-driven journalism.
Because of its multi-layered nature, science hype carries several disturbing consequences. For one, exaggerated coverage of a discovery could mislead the public by giving them false hope or unfounded worry. And media hype can contribute to a general mistrust of science. In these instances, people might, as Auch puts it, "fall back on previously held beliefs, evocative narratives, or comforting biases instead of well-justified scientific evidence."
All of this is especially dangerous in today's 'fake news' era, when companies or political parties sow public confusion for their own benefit, such as with global warming. In the case of Nectome, the danger is that people might opt to end their lives based off a lacking scientific theory. In fact, the company is hoping to enlist terminal patients in California, where doctor-assisted suicide is legal. And 25 people have paid the $10,000 to join Nectome's waiting list, including Sam Altman, president of the famed startup accelerator Y Combinator. Nectome now has offered to refund the money.
Founders McCanna and McIntyre did not return repeated requests for comment for this article. A new statement on their website begins: "Vitrifixation today is a powerful research tool, but needs more research and development before anyone considers applying it in a context other than research."
The solution to the dangers of hype, experts say, is a more scientifically literate public—and less clickbait-driven journalism. Until then, it seems that companies like Nectome will continue to enjoy at least 15 minutes of fame.
DNA- and RNA-based electronic implants may revolutionize healthcare
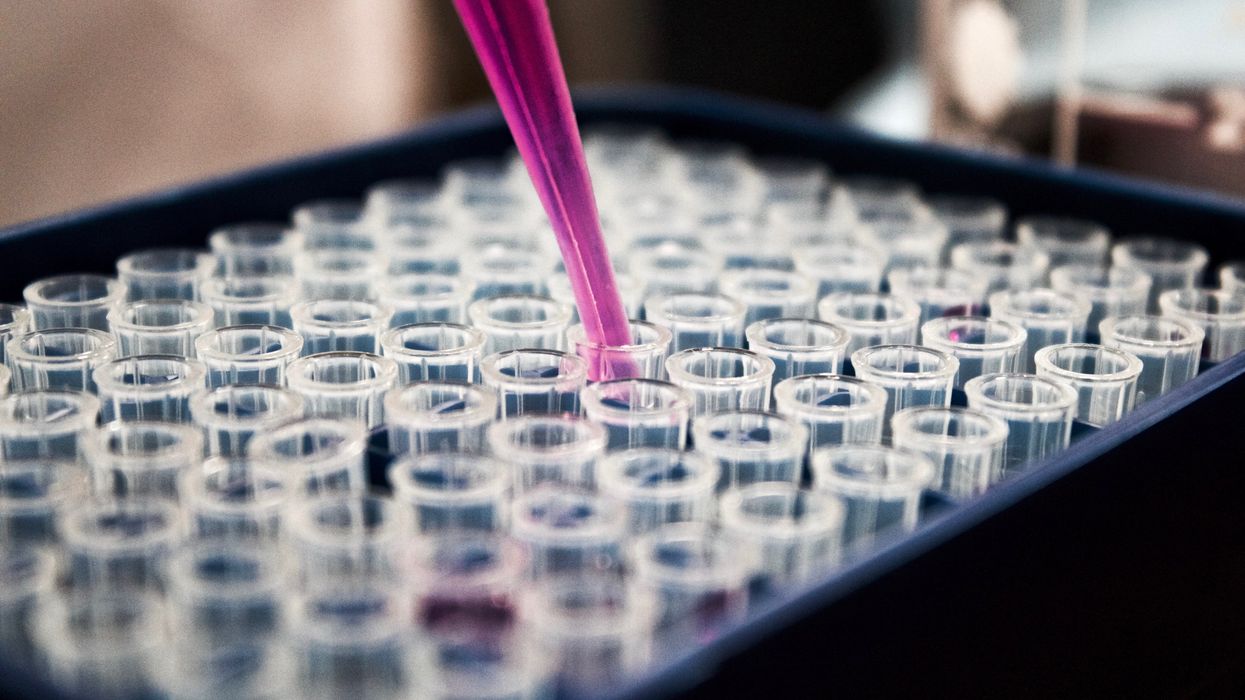
The test tubes contain tiny DNA/enzyme-based circuits, which comprise TRUMPET, a new type of electronic device, smaller than a cell.
Implantable electronic devices can significantly improve patients’ quality of life. A pacemaker can encourage the heart to beat more regularly. A neural implant, usually placed at the back of the skull, can help brain function and encourage higher neural activity. Current research on neural implants finds them helpful to patients with Parkinson’s disease, vision loss, hearing loss, and other nerve damage problems. Several of these implants, such as Elon Musk’s Neuralink, have already been approved by the FDA for human use.
Yet, pacemakers, neural implants, and other such electronic devices are not without problems. They require constant electricity, limited through batteries that need replacements. They also cause scarring. “The problem with doing this with electronics is that scar tissue forms,” explains Kate Adamala, an assistant professor of cell biology at the University of Minnesota Twin Cities. “Anytime you have something hard interacting with something soft [like muscle, skin, or tissue], the soft thing will scar. That's why there are no long-term neural implants right now.” To overcome these challenges, scientists are turning to biocomputing processes that use organic materials like DNA and RNA. Other promised benefits include “diagnostics and possibly therapeutic action, operating as nanorobots in living organisms,” writes Evgeny Katz, a professor of bioelectronics at Clarkson University, in his book DNA- And RNA-Based Computing Systems.
While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output.
Adamala’s research focuses on developing such biocomputing systems using DNA, RNA, proteins, and lipids. Using these molecules in the biocomputing systems allows the latter to be biocompatible with the human body, resulting in a natural healing process. In a recent Nature Communications study, Adamala and her team created a new biocomputing platform called TRUMPET (Transcriptional RNA Universal Multi-Purpose GatE PlaTform) which acts like a DNA-powered computer chip. “These biological systems can heal if you design them correctly,” adds Adamala. “So you can imagine a computer that will eventually heal itself.”
The basics of biocomputing
Biocomputing and regular computing have many similarities. Like regular computing, biocomputing works by running information through a series of gates, usually logic gates. A logic gate works as a fork in the road for an electronic circuit. The input will travel one way or another, giving two different outputs. An example logic gate is the AND gate, which has two inputs (A and B) and two different results. If both A and B are 1, the AND gate output will be 1. If only A is 1 and B is 0, the output will be 0 and vice versa. If both A and B are 0, the result will be 0. While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output. In this case, the DNA enters the logic gate as a single or double strand.
If the DNA is double-stranded, the system “digests” the DNA or destroys it, which results in non-fluorescence or “0” output. Conversely, if the DNA is single-stranded, it won’t be digested and instead will be copied by several enzymes in the biocomputing system, resulting in fluorescent RNA or a “1” output. And the output for this type of binary system can be expanded beyond fluorescence or not. For example, a “1” output might be the production of the enzyme insulin, while a “0” may be that no insulin is produced. “This kind of synergy between biology and computation is the essence of biocomputing,” says Stephanie Forrest, a professor and the director of the Biodesign Center for Biocomputing, Security and Society at Arizona State University.
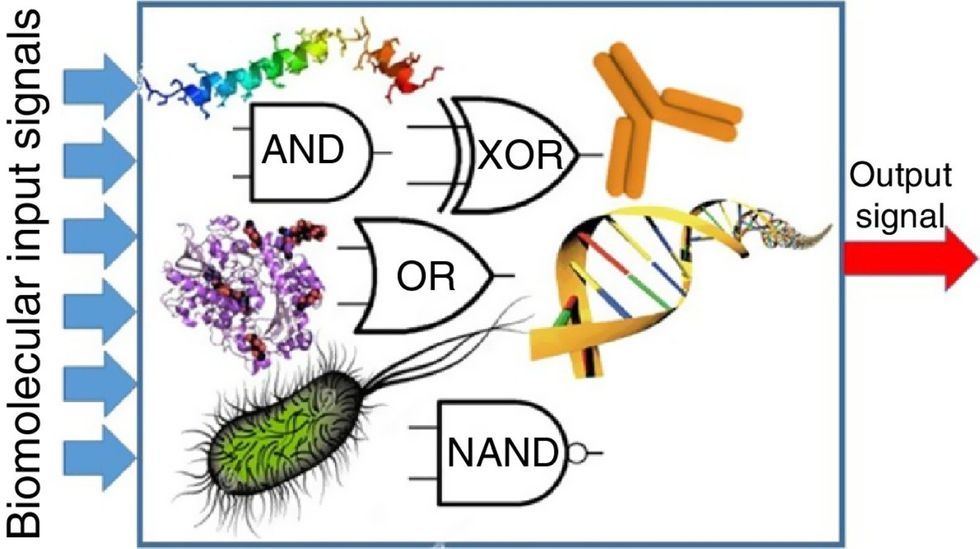
Biocomputing circles are made of DNA, RNA, proteins and even bacteria.
Evgeny Katz
The TRUMPET’s promise
Depending on whether the biocomputing system is placed directly inside a cell within the human body, or run in a test-tube, different environmental factors play a role. When an output is produced inside a cell, the cell's natural processes can amplify this output (for example, a specific protein or DNA strand), creating a solid signal. However, these cells can also be very leaky. “You want the cells to do the thing you ask them to do before they finish whatever their businesses, which is to grow, replicate, metabolize,” Adamala explains. “However, often the gate may be triggered without the right inputs, creating a false positive signal. So that's why natural logic gates are often leaky." While biocomputing outside a cell in a test tube can allow for tighter control over the logic gates, the outputs or signals cannot be amplified by a cell and are less potent.
TRUMPET, which is smaller than a cell, taps into both cellular and non-cellular biocomputing benefits. “At its core, it is a nonliving logic gate system,” Adamala states, “It's a DNA-based logic gate system. But because we use enzymes, and the readout is enzymatic [where an enzyme replicates the fluorescent RNA], we end up with signal amplification." This readout means that the output from the TRUMPET system, a fluorescent RNA strand, can be replicated by nearby enzymes in the platform, making the light signal stronger. "So it combines the best of both worlds,” Adamala adds.
These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body.
The TRUMPET biocomputing process is relatively straightforward. “If the DNA [input] shows up as single-stranded, it will not be digested [by the logic gate], and you get this nice fluorescent output as the RNA is made from the single-stranded DNA, and that's a 1,” Adamala explains. "And if the DNA input is double-stranded, it gets digested by the enzymes in the logic gate, and there is no RNA created from the DNA, so there is no fluorescence, and the output is 0." On the story's leading image above, if the tube is "lit" with a purple color, that is a binary 1 signal for computing. If it's "off" it is a 0.
While still in research, TRUMPET and other biocomputing systems promise significant benefits to personalized healthcare and medicine. These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body. The study’s lead author and graduate student Judee Sharon is already beginning to research TRUMPET's ability for earlier cancer diagnoses. Because the inputs for TRUMPET are single or double-stranded DNA, any mutated or cancerous DNA could theoretically be detected from the platform through the biocomputing process. Theoretically, devices like TRUMPET could be used to detect cancer and other diseases earlier.
Adamala sees TRUMPET not only as a detection system but also as a potential cancer drug delivery system. “Ideally, you would like the drug only to turn on when it senses the presence of a cancer cell. And that's how we use the logic gates, which work in response to inputs like cancerous DNA. Then the output can be the production of a small molecule or the release of a small molecule that can then go and kill what needs killing, in this case, a cancer cell. So we would like to develop applications that use this technology to control the logic gate response of a drug’s delivery to a cell.”
Although platforms like TRUMPET are making progress, a lot more work must be done before they can be used commercially. “The process of translating mechanisms and architecture from biology to computing and vice versa is still an art rather than a science,” says Forrest. “It requires deep computer science and biology knowledge,” she adds. “Some people have compared interdisciplinary science to fusion restaurants—not all combinations are successful, but when they are, the results are remarkable.”
Crickets are low on fat, high on protein, and can be farmed sustainably. They are also crunchy.
In today’s podcast episode, Leaps.org Deputy Editor Lina Zeldovich speaks about the health and ecological benefits of farming crickets for human consumption with Bicky Nguyen, who joins Lina from Vietnam. Bicky and her business partner Nam Dang operate an insect farm named CricketOne. Motivated by the idea of sustainable and healthy protein production, they started their unconventional endeavor a few years ago, despite numerous naysayers who didn’t believe that humans would ever consider munching on bugs.
Yet, making creepy crawlers part of our diet offers many health and planetary advantages. Food production needs to match the rise in global population, estimated to reach 10 billion by 2050. One challenge is that some of our current practices are inefficient, polluting and wasteful. According to nonprofit EarthSave.org, it takes 2,500 gallons of water, 12 pounds of grain, 35 pounds of topsoil and the energy equivalent of one gallon of gasoline to produce one pound of feedlot beef, although exact statistics vary between sources.
Meanwhile, insects are easy to grow, high on protein and low on fat. When roasted with salt, they make crunchy snacks. When chopped up, they transform into delicious pâtes, says Bicky, who invents her own cricket recipes and serves them at industry and public events. Maybe that’s why some research predicts that edible insects market may grow to almost $10 billion by 2030. Tune in for a delectable chat on this alternative and sustainable protein.
Listen on Apple | Listen on Spotify | Listen on Stitcher | Listen on Amazon | Listen on Google
Further reading:
More info on Bicky Nguyen
https://yseali.fulbright.edu.vn/en/faculty/bicky-n...
The environmental footprint of beef production
https://www.earthsave.org/environment.htm
https://www.watercalculator.org/news/articles/beef-king-big-water-footprints/
https://www.frontiersin.org/articles/10.3389/fsufs.2019.00005/full
https://ourworldindata.org/carbon-footprint-food-methane
Insect farming as a source of sustainable protein
https://www.insectgourmet.com/insect-farming-growing-bugs-for-protein/
https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/insect-farming
Cricket flour is taking the world by storm
https://www.cricketflours.com/
https://talk-commerce.com/blog/what-brands-use-cricket-flour-and-why/

Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.