Scientists search for a universal coronavirus vaccine

Stephen Zeichner, an infectious disease specialist at the University of Virginia Medical Center, has made progress with an early stage universal coronavirus vaccine.
The Covid-19 pandemic had barely begun when VBI Vaccines, a biopharmaceutical company based in Cambridge, Massachusetts, initiated their search for a universal coronavirus vaccine.
It was March 2020, and while most pharmaceutical companies were scrambling to initiate vaccine programs which specifically targeted the SARS-CoV-2 virus, VBI’s executives were already keen to look at the broader picture.
Having observed the SARS and MERS coronavirus outbreaks over the last two decades, Jeff Baxter, CEO of VBI Vaccines, was aware that SARS-CoV-2 is unlikely to be the last coronavirus to move from an animal host into humans. “It's absolutely apparent that the future is to create a vaccine which gives more broad protection against not only pre-existing coronaviruses, but those that will potentially make the leap into humans in future,” says Baxter.
It was a prescient decision. Over the last two years, more biotechs and pharma companies have joined the search to find a vaccine which might be able to protect against all coronaviruses, along with dozens of academic research groups. Last September, the US National Institutes of Health dedicated $36 million specifically to pan-coronavirus vaccine research, while the global Coalition for Epidemic Preparedness Innovations (CEPI) has earmarked $200 million towards the effort.
Until October 2021, the very concept of whether it might be
theoretically possible to vaccinate against multiple coronaviruses remained an open question. But then a groundbreaking study renewed optimism.
The emergence of new variants of Covid-19 over the past year, particularly the highly mutated Omicron variant, has added greater impetus to find broader spectrum vaccines. But until October 2021, the very concept of whether it might be theoretically possible to vaccinate against multiple coronaviruses remained an open question. After all, scientists have spent decades trying to develop a similar vaccine for influenza with little success.
But then a groundbreaking study from renowned virologist Linfa Wang, who runs the emerging infectious diseases program at Duke-National University of Singapore Medical School, provided renewed optimism.
Wang found that eight SARS survivors who had been injected with the Pfizer/BioNTech Covid-19 vaccine had neutralising antibodies in their blood against SARS, the Alpha, Beta and Delta variants of SARS-CoV-2, and five other coronaviruses which reside in bats and pangolins. He concluded that the combination of past coronavirus infection, and immunization with a messenger RNA vaccine, had resulted in a wider spectrum of protection than might have been expected.
“This is a significant study because it showed that pre-existing immunity to one coronavirus could help with the elicitation of cross-reactive antibodies when immunizing with a second coronavirus,” says Kevin Saunders, Director of Research at the Duke Human Vaccine Institute in North Carolina, which is developing a universal coronavirus vaccine. “It provides a strategy to perhaps broaden the immune response against coronaviruses.”
In the next few months, some of the first data is set to emerge looking at whether this kind of antibody response could be elicited by a single universal coronavirus vaccine. In April 2021, scientists at the Walter Reed Army Institute of Research in Silver Spring, Maryland, launched a Phase I clinical trial of their vaccine, with a spokesman saying that it was successful, and the full results will be announced soon.
The Walter Reed researchers have already released preclinical data, testing the vaccine in non-human primates where it was found to have immunising capabilities against a range of Covid-19 variants as well as the original SARS virus. If the Phase I trial displays similar efficacy, a larger Phase II trial will begin later this year.
Two different approaches
Broadly speaking, scientists are taking two contrasting approaches to the task of finding a universal coronavirus vaccine. The Walter Reed Army Institute of Research, VBI Vaccines – who plan to launch their own clinical trial in the summer – and the Duke Human Vaccine Institute – who are launching a Phase I trial in early 2023 – are using a soccer-ball shaped ferritin nanoparticle studded with different coronavirus protein fragments.
VBI Vaccines is looking to elicit broader immune responses by combining SARS, SARS-CoV-2 and MERS spike proteins on the same nanoparticle. Dave Anderson, chief scientific officer at VBI Vaccines, explains that the idea is that by showing the immune system these three spike proteins at the same time, it can help train it to identify and respond to subtle differences between coronavirus strains.
The Duke Human Vaccine Institute is utilising the same method, but rather than including the entire spike proteins from different coronaviruses, they are only including the receptor binding domain (RBD) fragment from each spike protein. “We designed our vaccine to focus the immune system on a site of vulnerability for the virus, which is the receptor binding domain,” says Saunders. “Since the RBD is small, arraying multiple RBDs on a nanoparticle is a straight-forward approach. The goal is to generate immunity to many different subgenuses of viruses so that there will be cross-reactivity with new or unknown coronaviruses.”
But the other strategy is to create a vaccine which contains regions of the viral protein structure which are conserved between all coronavirus strains. This is something which scientists have tried to do for a universal influenza vaccine, but it is thought to be more feasible for coronaviruses because they mutate at a slower rate and are more constrained in the ways that they can evolve.
DIOSynVax, a biotech based in Cambridge, United Kingdom, announced in a press release earlier this month that they are partnering with CEPI to use their computational predictive modelling techniques to identify common structures between all of the SARS coronaviruses which do not mutate, and thus present good vaccine targets.
Stephen Zeichner, an infectious disease specialist at the University of Virginia Medical Center, has created an early stage vaccine using the fusion peptide region – another part of the coronavirus spike protein that aids the virus’s entry into host cells – which so far appears to be highly conserved between all coronaviruses.
So far Zeichner has trialled this version of the vaccine in pigs, where it provided protection against a different coronavirus called porcine epidemic diarrhea virus, which he described as very promising as this virus is from a different family called alphacoronaviruses, while SARS-CoV-2 is a betacoronavirus.
“If a betacoronavirus fusion peptide vaccine designed from SARS-CoV-2 can protect pigs against clinical disease from an alphacoronavirus, then that suggests that an analogous vaccine would enable broad protection against many, many different coronaviruses,” he says.
The road ahead
But while some of the early stage results are promising, researchers are fully aware of the scale of the challenge ahead of them. Although CEPI have declared an aim of having a licensed universal coronavirus vaccine available by 2024-2025, Zeichner says that such timelines are ambitious in the extreme.
“I was incredibly impressed at the speed at which the mRNA coronavirus vaccines were developed for SARS-CoV-2,” he says. “That was faster than just about anybody anticipated. On the other hand, I think a universal coronavirus vaccine is more equivalent to the challenge of developing an HIV vaccine and we're 35 years into that effort without success. We know a lot more now than before, and maybe it will be easier than we think. But I think the route to a universal vaccine is harder than an individual vaccine, so I wouldn’t want to put money on a timeline prediction.”
The major challenge for scientists is essentially designing a vaccine for a future threat which is not even here yet. As such, there are no guidelines on what safety data would be required to license such a vaccine, and how researchers can demonstrate that it truly provides efficacy against all coronaviruses, even those which have not yet jumped to humans.
The teams working on this problem have already devised some ingenious ways of approaching the challenge. VBI Vaccines have taken the genetic sequences of different coronaviruses found in bats and pangolins, from publicly available databases, and inserted them into what virologists call a pseudotype virus – one which has been engineered so it does not have enough genetic material to replicate.
This has allowed them to test the neutralising antibodies that their vaccine produces against these coronaviruses in test tubes, under safe lab conditions. “We have literally just been ordering the sequences, and making synthetic viruses that we can use to test the antibody responses,” says Anderson.
However, some scientists feel that going straight to a universal coronavirus vaccine is likely to be too complex. Instead they say that we should aim for vaccines which are a little more specific. Pamela Bjorkman, a structural biologist at the California Institute of Technology, suggests that pan-coronavirus vaccines which protect against SARS-like betacoronaviruses such as SARS or SARS-CoV-2, or MERS-like betacoronaviruses, may be more realistic.
“I think a vaccine to protect against all coronaviruses is likely impossible since there are so many varieties,” she says. “Perhaps trying to narrow down the scope is advisable.”
But if the mission to develop a universal coronavirus vaccine does succeed, it will be one of the most remarkable feats in the annals of medical science. In January, US chief medical advisor Anthony Fauci urged for greater efforts to be devoted towards this goal, one which scientists feel would be the biological equivalent of the race to develop the first atomic bomb
“The development of an effective universal coronavirus vaccine would be equally groundbreaking, as it would have global applicability and utility,” says Saunders. “Coronaviruses have caused multiple deadly outbreaks, and it is likely that another outbreak will occur. Having a vaccine that prevents death from a future outbreak would be a tremendous achievement in global health.”
He agrees that it will require creativity on a remarkable scale: “The universal coronavirus vaccine will also require ingenuity and perseverance comparable to that needed for the Manhattan project.”
DNA- and RNA-based electronic implants may revolutionize healthcare
The test tubes contain tiny DNA/enzyme-based circuits, which comprise TRUMPET, a new type of electronic device, smaller than a cell.
Implantable electronic devices can significantly improve patients’ quality of life. A pacemaker can encourage the heart to beat more regularly. A neural implant, usually placed at the back of the skull, can help brain function and encourage higher neural activity. Current research on neural implants finds them helpful to patients with Parkinson’s disease, vision loss, hearing loss, and other nerve damage problems. Several of these implants, such as Elon Musk’s Neuralink, have already been approved by the FDA for human use.
Yet, pacemakers, neural implants, and other such electronic devices are not without problems. They require constant electricity, limited through batteries that need replacements. They also cause scarring. “The problem with doing this with electronics is that scar tissue forms,” explains Kate Adamala, an assistant professor of cell biology at the University of Minnesota Twin Cities. “Anytime you have something hard interacting with something soft [like muscle, skin, or tissue], the soft thing will scar. That's why there are no long-term neural implants right now.” To overcome these challenges, scientists are turning to biocomputing processes that use organic materials like DNA and RNA. Other promised benefits include “diagnostics and possibly therapeutic action, operating as nanorobots in living organisms,” writes Evgeny Katz, a professor of bioelectronics at Clarkson University, in his book DNA- And RNA-Based Computing Systems.
While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output.
Adamala’s research focuses on developing such biocomputing systems using DNA, RNA, proteins, and lipids. Using these molecules in the biocomputing systems allows the latter to be biocompatible with the human body, resulting in a natural healing process. In a recent Nature Communications study, Adamala and her team created a new biocomputing platform called TRUMPET (Transcriptional RNA Universal Multi-Purpose GatE PlaTform) which acts like a DNA-powered computer chip. “These biological systems can heal if you design them correctly,” adds Adamala. “So you can imagine a computer that will eventually heal itself.”
The basics of biocomputing
Biocomputing and regular computing have many similarities. Like regular computing, biocomputing works by running information through a series of gates, usually logic gates. A logic gate works as a fork in the road for an electronic circuit. The input will travel one way or another, giving two different outputs. An example logic gate is the AND gate, which has two inputs (A and B) and two different results. If both A and B are 1, the AND gate output will be 1. If only A is 1 and B is 0, the output will be 0 and vice versa. If both A and B are 0, the result will be 0. While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output. In this case, the DNA enters the logic gate as a single or double strand.
If the DNA is double-stranded, the system “digests” the DNA or destroys it, which results in non-fluorescence or “0” output. Conversely, if the DNA is single-stranded, it won’t be digested and instead will be copied by several enzymes in the biocomputing system, resulting in fluorescent RNA or a “1” output. And the output for this type of binary system can be expanded beyond fluorescence or not. For example, a “1” output might be the production of the enzyme insulin, while a “0” may be that no insulin is produced. “This kind of synergy between biology and computation is the essence of biocomputing,” says Stephanie Forrest, a professor and the director of the Biodesign Center for Biocomputing, Security and Society at Arizona State University.
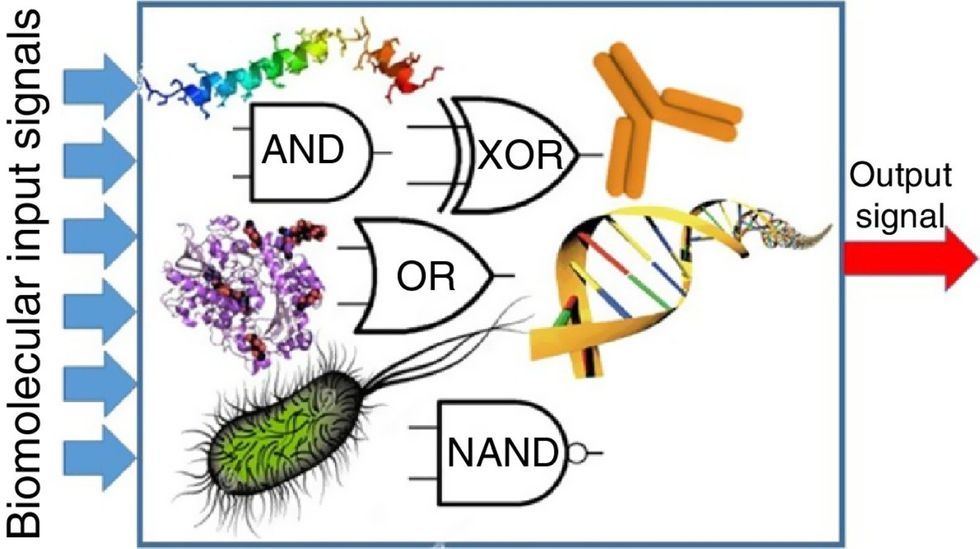
Biocomputing circles are made of DNA, RNA, proteins and even bacteria.
Evgeny Katz
The TRUMPET’s promise
Depending on whether the biocomputing system is placed directly inside a cell within the human body, or run in a test-tube, different environmental factors play a role. When an output is produced inside a cell, the cell's natural processes can amplify this output (for example, a specific protein or DNA strand), creating a solid signal. However, these cells can also be very leaky. “You want the cells to do the thing you ask them to do before they finish whatever their businesses, which is to grow, replicate, metabolize,” Adamala explains. “However, often the gate may be triggered without the right inputs, creating a false positive signal. So that's why natural logic gates are often leaky." While biocomputing outside a cell in a test tube can allow for tighter control over the logic gates, the outputs or signals cannot be amplified by a cell and are less potent.
TRUMPET, which is smaller than a cell, taps into both cellular and non-cellular biocomputing benefits. “At its core, it is a nonliving logic gate system,” Adamala states, “It's a DNA-based logic gate system. But because we use enzymes, and the readout is enzymatic [where an enzyme replicates the fluorescent RNA], we end up with signal amplification." This readout means that the output from the TRUMPET system, a fluorescent RNA strand, can be replicated by nearby enzymes in the platform, making the light signal stronger. "So it combines the best of both worlds,” Adamala adds.
These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body.
The TRUMPET biocomputing process is relatively straightforward. “If the DNA [input] shows up as single-stranded, it will not be digested [by the logic gate], and you get this nice fluorescent output as the RNA is made from the single-stranded DNA, and that's a 1,” Adamala explains. "And if the DNA input is double-stranded, it gets digested by the enzymes in the logic gate, and there is no RNA created from the DNA, so there is no fluorescence, and the output is 0." On the story's leading image above, if the tube is "lit" with a purple color, that is a binary 1 signal for computing. If it's "off" it is a 0.
While still in research, TRUMPET and other biocomputing systems promise significant benefits to personalized healthcare and medicine. These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body. The study’s lead author and graduate student Judee Sharon is already beginning to research TRUMPET's ability for earlier cancer diagnoses. Because the inputs for TRUMPET are single or double-stranded DNA, any mutated or cancerous DNA could theoretically be detected from the platform through the biocomputing process. Theoretically, devices like TRUMPET could be used to detect cancer and other diseases earlier.
Adamala sees TRUMPET not only as a detection system but also as a potential cancer drug delivery system. “Ideally, you would like the drug only to turn on when it senses the presence of a cancer cell. And that's how we use the logic gates, which work in response to inputs like cancerous DNA. Then the output can be the production of a small molecule or the release of a small molecule that can then go and kill what needs killing, in this case, a cancer cell. So we would like to develop applications that use this technology to control the logic gate response of a drug’s delivery to a cell.”
Although platforms like TRUMPET are making progress, a lot more work must be done before they can be used commercially. “The process of translating mechanisms and architecture from biology to computing and vice versa is still an art rather than a science,” says Forrest. “It requires deep computer science and biology knowledge,” she adds. “Some people have compared interdisciplinary science to fusion restaurants—not all combinations are successful, but when they are, the results are remarkable.”
Crickets are low on fat, high on protein, and can be farmed sustainably. They are also crunchy.
In today’s podcast episode, Leaps.org Deputy Editor Lina Zeldovich speaks about the health and ecological benefits of farming crickets for human consumption with Bicky Nguyen, who joins Lina from Vietnam. Bicky and her business partner Nam Dang operate an insect farm named CricketOne. Motivated by the idea of sustainable and healthy protein production, they started their unconventional endeavor a few years ago, despite numerous naysayers who didn’t believe that humans would ever consider munching on bugs.
Yet, making creepy crawlers part of our diet offers many health and planetary advantages. Food production needs to match the rise in global population, estimated to reach 10 billion by 2050. One challenge is that some of our current practices are inefficient, polluting and wasteful. According to nonprofit EarthSave.org, it takes 2,500 gallons of water, 12 pounds of grain, 35 pounds of topsoil and the energy equivalent of one gallon of gasoline to produce one pound of feedlot beef, although exact statistics vary between sources.
Meanwhile, insects are easy to grow, high on protein and low on fat. When roasted with salt, they make crunchy snacks. When chopped up, they transform into delicious pâtes, says Bicky, who invents her own cricket recipes and serves them at industry and public events. Maybe that’s why some research predicts that edible insects market may grow to almost $10 billion by 2030. Tune in for a delectable chat on this alternative and sustainable protein.
Listen on Apple | Listen on Spotify | Listen on Stitcher | Listen on Amazon | Listen on Google
Further reading:
More info on Bicky Nguyen
https://yseali.fulbright.edu.vn/en/faculty/bicky-n...
The environmental footprint of beef production
https://www.earthsave.org/environment.htm
https://www.watercalculator.org/news/articles/beef-king-big-water-footprints/
https://www.frontiersin.org/articles/10.3389/fsufs.2019.00005/full
https://ourworldindata.org/carbon-footprint-food-methane
Insect farming as a source of sustainable protein
https://www.insectgourmet.com/insect-farming-growing-bugs-for-protein/
https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/insect-farming
Cricket flour is taking the world by storm
https://www.cricketflours.com/
https://talk-commerce.com/blog/what-brands-use-cricket-flour-and-why/

Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.