When doctors couldn’t stop her daughter’s seizures, this mom earned a PhD and found a treatment herself.

Savannah Salazar (left) and her mother, Tracy Dixon-Salazaar, who earned a PhD in neurobiology in the quest for a treatment of her daughter's seizure disorder.
Twenty-eight years ago, Tracy Dixon-Salazaar woke to the sound of her daughter, two-year-old Savannah, in the midst of a medical emergency.
“I entered [Savannah’s room] to see her tiny little body jerking about violently in her bed,” Tracy said in an interview. “I thought she was choking.” When she and her husband frantically called 911, the paramedic told them it was likely that Savannah had had a seizure—a term neither Tracy nor her husband had ever heard before.
Over the next several years, Savannah’s seizures continued and worsened. By age five Savannah was having seizures dozens of times each day, and her parents noticed significant developmental delays. Savannah was unable to use the restroom and functioned more like a toddler than a five-year-old.
Doctors were mystified: Tracy and her husband had no family history of seizures, and there was no event—such as an injury or infection—that could have caused them. Doctors were also confused as to why Savannah’s seizures were happening so frequently despite trying different seizure medications.
Doctors eventually diagnosed Savannah with Lennox-Gaustaut Syndrome, or LGS, an epilepsy disorder with no cure and a poor prognosis. People with LGS are often resistant to several kinds of anti-seizure medications, and often suffer from developmental delays and behavioral problems. People with LGS also have a higher chance of injury as well as a higher chance of sudden unexpected death (SUDEP) due to the frequent seizures. In about 70 percent of cases, LGS has an identifiable cause such as a brain injury or genetic syndrome. In about 30 percent of cases, however, the cause is unknown.
Watching her daughter struggle through repeated seizures was devastating to Tracy and the rest of the family.
“This disease, it comes into your life. It’s uninvited. It’s unannounced and it takes over every aspect of your daily life,” said Tracy in an interview with Today.com. “Plus it’s attacking the thing that is most precious to you—your kid.”
Desperate to find some answers, Tracy began combing the medical literature for information about epilepsy and LGS. She enrolled in college courses to better understand the papers she was reading.
“Ironically, I thought I needed to go to college to take English classes to understand these papers—but soon learned it wasn’t English classes I needed, It was science,” Tracy said. When she took her first college science course, Tracy says, she “fell in love with the subject.”
Tracy was now a caregiver to Savannah, who continued to have hundreds of seizures a month, as well as a full-time student, studying late into the night and while her kids were at school, using classwork as “an outlet for the pain.”
“I couldn’t help my daughter,” Tracy said. “Studying was something I could do.”
Twelve years later, Tracy had earned a PhD in neurobiology.
After her post-doctoral training, Tracy started working at a lab that explored the genetics of epilepsy. Savannah’s doctors hadn’t found a genetic cause for her seizures, so Tracy decided to sequence her genome again to check for other abnormalities—and what she found was life-changing.
Tracy discovered that Savannah had a calcium channel mutation, meaning that too much calcium was passing through Savannah’s neural pathways, leading to seizures. The information made sense to Tracy: Anti-seizure medications often leech calcium from a person’s bones. When doctors had prescribed Savannah calcium supplements in the past to counteract these effects, her seizures had gotten worse every time she took the medication. Tracy took her discovery to Savannah’s doctor, who agreed to prescribe her a calcium blocker.
The change in Savannah was almost immediate.
Within two weeks, Savannah’s seizures had decreased by 95 percent. Once on a daily seven-drug regimen, she was soon weaned to just four, and then three. Amazingly, Tracy started to notice changes in Savannah’s personality and development, too.
“She just exploded in her personality and her talking and her walking and her potty training and oh my gosh she is just so sassy,” Tracy said in an interview.
Since starting the calcium blocker eleven years ago, Savannah has continued to make enormous strides. Though still unable to read or write, Savannah enjoys puzzles and social media. She’s “obsessed” with boys, says Tracy. And while Tracy suspects she’ll never be able to live independently, she and her daughter can now share more “normal” moments—something she never anticipated at the start of Savannah’s journey with LGS. While preparing for an event, Savannah helped Tracy get ready.
“We picked out a dress and it was the first time in our lives that we did something normal as a mother and a daughter,” she said. “It was pretty cool.”
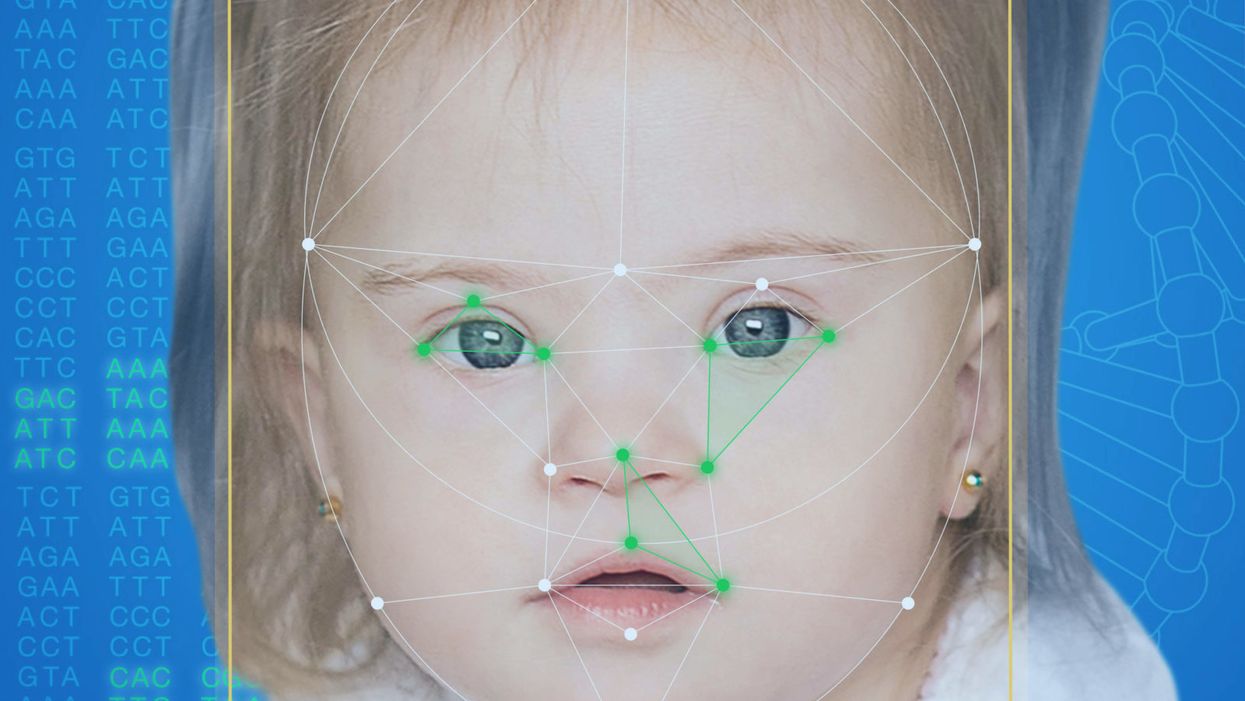
This App Helps Diagnose Rare Genetic Disorders from a Picture
FDNA's Face2Gene technology analyzes patient biometric data using artificial intelligence, identifying correlations with disease-causing genetic variations.
Medical geneticist Omar Abdul-Rahman had a hunch. He thought that the three-year-old boy with deep-set eyes, a rounded nose, and uplifted earlobes might have Mowat-Wilson syndrome, but he'd never seen a patient with the rare disorder before.
"If it weren't for the app I'm not sure I would have had the confidence to say 'yes you should spend $1000 on this test."
Rahman had already ordered genetic tests for three different conditions without any luck, and he didn't want to cost the family any more money—or hope—if he wasn't sure of the diagnosis. So he took a picture of the boy and uploaded the photo to Face2Gene, a diagnostic aid for rare genetic disorders. Sure enough, Mowat-Wilson came up as a potential match. The family agreed to one final genetic test, which was positive for the syndrome.
"If it weren't for the app I'm not sure I would have had the confidence to say 'yes you should spend $1000 on this test,'" says Rahman, who is now the director of Genetic Medicine at the University of Nebraska Medical Center, but saw the boy when he was in the Department of Pediatrics at the University of Mississippi Medical Center in 2012.
"Families who are dealing with undiagnosed diseases never know what's going to come around the corner, what other organ system might be a problem next week," Rahman says. With a diagnosis, "You don't have to wait for the other shoe to drop because now you know the extent of the condition."
A diagnosis is the first and most important step for patients to attain medical care. Disease prognosis, treatment plans, and emotional coping all stem from this critical phase. But diagnosis can also be the trickiest part of the process, particularly for rare disorders. According to one European survey, 40 percent of rare diseases are initially misdiagnosed.
Healthcare professionals and medical technology companies hope that facial recognition software will help prevent families from facing difficult disruptions due to misdiagnoses.
"Patients with rare diseases or genetic disorders go through a long period of diagnostic odyssey, and just putting a name to a syndrome or finding a diagnosis can be very helpful and relieve a lot of tension for the family," says Dekel Gelbman, CEO of FDNA.
Consequently, a misdiagnosis can be devastating for families. Money and time may have been wasted on fruitless treatments, while opportunities for potentially helpful therapies or clinical trials were missed. Parents led down the wrong path must change their expectations of their child's long-term prognosis and care. In addition, they may be misinformed regarding future decisions about family planning.
Healthcare professionals and medical technology companies hope that facial recognition software will help prevent families from facing these difficult disruptions by improving the accuracy and ease of diagnosing genetic disorders. Traditionally, doctors diagnose these types of conditions by identifying unique patterns of facial features, a practice called dysmorphology. Trained physicians can read a child's face like a map and detect any abnormal ridges or plateaus—wide-set eyes, broad forehead, flat nose, rotated ears—that, combined with other symptoms such as intellectual disability or abnormal height and weight, signify a specific genetic disorder.
These morphological changes can be subtle, though, and often only specialized medical geneticists are able to detect and interpret these facial clues. What's more, some genetic disorders are so rare that even a specialist may not have encountered it before, much less a general practitioner. Diagnosing rare conditions has improved thanks to genomic testing that can confirm (or refute) a doctor's suspicion. Yet with thousands of variants in each person's genome, identifying the culprit mutation or deletion can be extremely difficult if you don't know what you're looking for.
Facial recognition technology is trying to take some of the guesswork out of this process. Software such as the Face2Gene app use machine learning to compare a picture of a patient against images of thousands of disorders and come back with suggestions of possible diagnoses.
"This is a classic field for artificial intelligence because no human being can really have enough knowledge and enough experience to be able to do this for thousands of different disorders."
"When we met a geneticist for the first time we were pretty blown away with the fact that they actually use their own human pattern recognition" to diagnose patients, says Gelbman. "This is a classic field for AI [artificial intelligence], for machine learning because no human being can really have enough knowledge and enough experience to be able to do this for thousands of different disorders."
When a physician uploads a photo to the app, they are given a list of different diagnostic suggestions, each with a heat map to indicate how similar the facial features are to a classic representation of the syndrome. The physician can hone the suggestions by adding in other symptoms or family history. Gelbman emphasized that the app is a "search and reference tool" and should not "be used to diagnose or treat medical conditions." It is not approved by the FDA as a diagnostic.
"As a tool, we've all been waiting for this, something that can help everyone," says Julian Martinez-Agosto, an associate professor in human genetics and pediatrics at UCLA. He sees the greatest benefit of facial recognition technology in its ability to empower non-specialists to make a diagnosis. Many areas, including rural communities or resource-poor countries, do not have access to either medical geneticists trained in these types of diagnostics or genomic screens. Apps like Face2Gene can help guide a general practitioner or flag diseases they might not be familiar with.
One concern is that most textbook images of genetic disorders come from the West, so the "classic" face of a condition is often a child of European descent.
Maximilian Muenke, a senior investigator at the National Human Genome Research Institute (NHGRI), agrees that in many countries, facial recognition programs could be the only way for a doctor to make a diagnosis.
"There are only geneticists in countries like the U.S., Canada, Europe, Japan. In most countries, geneticists don't exist at all," Muenke says. "In Nigeria, the most populous country in all of Africa with 160 million people, there's not a single clinical geneticist. So in a country like that, facial recognition programs will be sought after and will be extremely useful to help make a diagnosis to the non-geneticists."
One concern about providing this type of technology to a global population is that most textbook images of genetic disorders come from the West, so the "classic" face of a condition is often a child of European descent. However, the defining facial features of some of these disorders manifest differently across ethnicities, leaving clinicians from other geographic regions at a disadvantage.
"Every syndrome is either more easy or more difficult to detect in people from different geographic backgrounds," explains Muenke. For example, "in some countries of Southeast Asia, the eyes are slanted upward, and that happens to be one of the findings that occurs mostly with children with Down Syndrome. So then it might be more difficult for some individuals to recognize Down Syndrome in children from Southeast Asia."
There is a risk that providing this type of diagnostic information online will lead to parents trying to classify their own children.
To combat this issue, Muenke helped develop the Atlas of Human Malformation Syndromes, a database that incorporates descriptions and pictures of patients from every continent. By providing examples of rare genetic disorders in children from outside of the United States and Europe, Muenke hopes to provide clinicians with a better understanding of what to look for in each condition, regardless of where they practice.
There is a risk that providing this type of diagnostic information online will lead to parents trying to classify their own children. Face2Gene is free to download in the app store, although users must be authenticated by the company as a healthcare professional before they can access the database. The NHGRI Atlas can be accessed by anyone through their website. However, Martinez and Muenke say parents already use Google and WebMD to look up their child's symptoms; facial recognition programs and databases are just an extension of that trend. In fact, Martinez says, "Empowering families is another way to facilitate access to care. Some families live in rural areas and have no access to geneticists. If they can use software to get a diagnosis and then contact someone at a large hospital, it can help facilitate the process."
Martinez also says the app could go further by providing greater transparency about how the program makes its assessments. Giving clinicians feedback about why a diagnosis fits certain facial features would offer a valuable teaching opportunity in addition to a diagnostic aid.
Both Martinez and Muenke think the technology is an innovation that could vastly benefit patients. "In the beginning, I was quite skeptical and I could not believe that a machine could replace a human," says Muenke. "However, I am a convert that it actually can help tremendously in making a diagnosis. I think there is a place for facial recognition programs, and I am a firm believer that this will spread over the next five years."
A thinking person.
"A world where people are slotted according to their inborn ability – well, that is Gattaca. That is eugenics."
This was the assessment of Dr. Catherine Bliss, a sociologist who wrote a new book on social science genetics, when asked by MIT Technology Review about polygenic scores that can predict a person's intelligence or performance in school. Like a credit score, a polygenic score is statistical tool that combines a lot of information about a person's genome into a single number. Fears about using polygenic scores for genetic discrimination are understandable, given this country's ugly history of using the science of heredity to justify atrocities like forcible sterilization. But polygenic scores are not the new eugenics. And, rushing to discuss polygenic scores in dystopian terms only contributes to widespread public misunderstanding about genetics.
Can we start genotyping toddlers to identify the budding geniuses among them? The short answer is no.
Let's begin with some background on how polygenic scores are developed. In a genome wide-association study, researchers conduct millions of statistical tests to identify small differences in people's DNA sequence that are correlated with differences in a target outcome (beyond what can attributed to chance or ancestry differences). Successful studies of this sort require enormous sample sizes, but companies like 23andMe are now contributing genetic data from their consumers to research studies, and national biorepositories like U.K. Biobank have put genetic information from hundreds of thousands of people online. When applied to studying blood lipids or myopia, this kind of study strikes people as a straightforward and uncontroversial scientific tool. But it can also be conducted for cognitive and behavioral outcomes, like how many years of school a person has completed. When researchers have finished a genome-wide association study, they are left with a dataset with millions of rows (one for each genetic variant analyzed) and one column with the correlations between each variant and the outcome being studied.
The trick to polygenic scoring is to use these results and apply them to people who weren't participants in the original study. Measure the genes of a new person, weight each one of her millions of genetic variants by its correlation with educational attainment from a genome-wide association study, and then simply add everything up into a single number. Voila! -- you've created a polygenic score for educational attainment. On its face, the idea of "scoring" a person's genotype does immediately suggest Gattaca-type applications. Can we now start screening embryos for their "inborn ability," as Bliss called it? Can we start genotyping toddlers to identify the budding geniuses among them?
The short answer is no. Here are four reasons why dystopian projections about polygenic scores are out of touch with the current science:
The phrase "DNA tests for IQ" makes for an attention-grabbing headline, but it's scientifically meaningless.
First, a polygenic score currently predicts the life outcomes of an individual child with a great deal of uncertainty. The amount of uncertainty around polygenic predictions will decrease in the future, as genetic discovery samples get bigger and genetic studies include more of the variation in the genome, including rare variants that are particular to a few families. But for now, knowing a child's polygenic score predicts his ultimate educational attainment about as well as knowing his family's income, and slightly worse than knowing how far his mother went in school. These pieces of information are also readily available about children before they are born, but no one is writing breathless think-pieces about the dystopian outcomes that will result from knowing whether a pregnant woman graduated from college.
Second, using polygenic scoring for embryo selection requires parents to create embryos using reproductive technology, rather than conceiving them by having sex. The prediction that many women will endure medically-unnecessary IVF, in order to select the embryo with the highest polygenic score, glosses over the invasiveness, indignity, pain, and heartbreak that these hormonal and surgical procedures can entail.
Third, and counterintuitively, a polygenic score might be using DNA to measure aspects of the child's environment. Remember, a child inherits her DNA from her parents, who typically also shape the environment she grows up in. And, children's environments respond to their unique personalities and temperaments. One Icelandic study found that parents' polygenic scores predicted their children's educational attainment, even if the score was constructed using only the half of the parental genome that the child didn't inherit. For example, imagine mom has genetic variant X that makes her more likely to smoke during her pregnancy. Prenatal exposure to nicotine, in turn, affects the child's neurodevelopment, leading to behavior problems in school. The school responds to his behavioral problems with suspension, causing him to miss out on instructional content. A genome-wide association study will collapse this long and winding causal path into a simple correlation -- "genetic variant X is correlated with academic achievement." But, a child's polygenic score, which includes variant X, will partly reflect his likelihood of being exposed to adverse prenatal and school environments.
Finally, the phrase "DNA tests for IQ" makes for an attention-grabbing headline, but it's scientifically meaningless. As I've written previously, it makes sense to talk about a bacterial test for strep throat, because strep throat is a medical condition defined as having streptococcal bacteria growing in the back of your throat. If your strep test is positive, you have strep throat, no matter how serious your symptoms are. But a polygenic score is not a test "for" IQ, because intelligence is not defined at the level of someone's DNA. It doesn't matter how high your polygenic score is, if you can't reason abstractly or learn from experience. Equating your intelligence, a cognitive capacity that is tested behaviorally, with your polygenic score, a number that is a weighted sum of genetic variants discovered to be statistically associated with educational attainment in a hypothesis-free data mining exercise, is misleading about what intelligence is and is not.
The task for many scientists like me, who are interested in understanding why some children do better in school than other children, is to disentangle correlations from causation.
So, if we're not going to build a Gattaca-style genetic hierarchy, what are polygenic scores good for? They are not useless. In fact, they give scientists a valuable new tool for studying how to improve children's lives. The task for many scientists like me, who are interested in understanding why some children do better in school than other children, is to disentangle correlations from causation. The best way to do that is to run an experiment where children are randomized to environments, but often a true experiment is unethical or impractical. You can't randomize children to be born to a teenage mother or to go to school with inexperienced teachers. By statistically controlling for some of the relevant genetic differences between people using a polygenic score, scientists are better able to identify potential environmental causes of differences in children's life outcomes. As we have seen with other methods from genetics, like twin studies, understanding genes illuminates the environment.
Research that examines genetics in relation to social inequality, such as differences in higher education outcomes, will obviously remind people of the horrors of the eugenics movement. Wariness regarding how genetic science will be applied is certainly warranted. But, polygenic scores are not pure measures of "inborn ability," and genome-wide association studies of human intelligence and educational attainment are not inevitably ushering in a new eugenics age.