“Young Blood” Transfusions Are Not Ready For Primetime – Yet
A young woman donates blood.
The world of dementia research erupted into cheers when news of the first real victory in a clinical trial against Alzheimer's Disease in over a decade was revealed last October.
By connecting the circulatory systems of a young and an old mouse, the regenerative potential of the young mouse decreased, and the old mouse became healthier.
Alzheimer's treatments have been famously difficult to develop; 99 percent of the 200-plus such clinical trials since 2000 have utterly failed. Even the few slight successes have failed to produce what is called 'disease modifying' agents that really help people with the disease. This makes the success, by the midsize Spanish pharma company Grifols, worthy of special attention.
However, the specifics of the Grifols treatment, a process called plasmapheresis, are atypical for another reason - they did not give patients a small molecule or an elaborate gene therapy, but rather simply the most common component of normal human blood plasma, a protein called albumin. A large portion of the patients' normal plasma was removed, and then a sterile solution of albumin was infused back into them to keep their overall blood volume relatively constant.
So why does replacing Alzheimer's patients' plasma with albumin seem to help their brains? One theory is that the action is direct. Alzheimer's patients have low levels of serum albumin, which is needed to clear out the plaques of amyloid that slowly build up in the brain. Supplementing those patients with extra albumin boosts their ability to clear the plaques and improves brain health. However, there is also evidence suggesting that the problem may be something present in the plasma of the sick person and pulling their plasma out and replacing it with a filler, like an albumin solution, may be what creates the purported benefit.
This scientific question is the tip of an iceberg that goes far beyond Alzheimer's Disease and albumin, to a debate that has been waged on the pages of scientific journals about the secrets of using young, healthy blood to extend youth and health.
This debate started long before the Grifols data was released, in 2014 when a group of researchers at Stanford found that by connecting the circulatory systems of a young and an old mouse, the regenerative potential of the young mouse decreased, and the old mouse became healthier. There was something either present in young blood that allowed tissues to regenerate, or something present in old blood that prevented regeneration. Whatever the biological reason, the effects in the experiment were extraordinary, providing a startling boost in health in the older mouse.
After the initial findings, multiple research groups got to work trying to identify the "active factor" of regeneration (or the inhibitor of that regeneration). They soon uncovered a variety of compounds such as insulin-like growth factor 1 (IGF1), CCL11, and GDF11, but none seemed to provide all the answers researchers were hoping for, with a number of high-profile retractions based on unsound experimental practices, or inconclusive data.
Years of research later, the simplest conclusion is that the story of plasma regeneration is not simple - there isn't a switch in our blood we can flip to turn back our biological clocks. That said, these hypotheses are far from dead, and many researchers continue to explore the possibility of using the rejuvenating ability of youthful plasma to treat a variety of diseases of aging.
But the bold claims of improved vigor thanks to young blood are so far unsupported by clinical evidence.
The data remain intriguing because of the astounding results from the conjoined circulatory system experiments. The current surge in interest in studying the biology of aging is likely to produce a new crop of interesting results in the next few years. Both CCL11 and GDF11 are being researched as potential drug targets by two startups, Alkahest and Elevian, respectively.
Without clarity on a single active factor driving rejuvenation, it's tempting to try a simpler approach: taking actual blood plasma provided by young people and infusing it into elderly subjects. This is what at least one startup company, Ambrosia, is now offering in five commercial clinics across the U.S. -- for $8,000 a liter.
By using whole plasma, the idea is to sidestep our ignorance, reaping the benefits of young plasma transfusion without knowing exactly what the active factors are that make the treatment work in mice. This space has attracted both established players in the plasmapheresis field – Alkahest and Grifols have teamed up to test fractions of whole plasma in Alzheimer's and Parkinson's – but also direct-to-consumer operations like Ambrosia that just want to offer patients access to treatments without regulatory oversight.
But the bold claims of improved vigor thanks to young blood are so far unsupported by clinical evidence. We simply haven't performed trials to test whether dosing a mostly healthy person with plasma can slow down aging, at least not yet. There is some evidence that plasma replacement works in mice, yes, but those experiments are all done in very different systems than what a human receiving young plasma might experience. To date, I have not seen any plasma transfusion clinic doing young blood plasmapheresis propose a clinical trial that is anything more than a shallow advertisement for their procedures.
The efforts I have seen to perform prophylactic plasmapheresis will fail to impact societal health. Without clearly defined endpoints and proper clinical trials, we won't know whether the procedure really lowers the risk of disease or helps with conditions of aging. So even if their hypothesis is correct, the lack of strong evidence to fall back on means that the procedure will never spread beyond the fringe groups willing to take the risk. If their hypothesis is wrong, then people are paying a huge amount of money for false hope, just as they do, sadly, at the phony stem cell clinics that started popping up all through the 2000s when stem cell hype was at its peak.
Until then, prophylactic plasma transfusions will be the domain of the optimistic and the gullible.
The real progress in the field will be made slowly, using carefully defined products either directly isolated from blood or targeting a bloodborne factor, just as the serious pharma and biotech players are doing already.
The field will progress in stages, first creating and carefully testing treatments for well-defined diseases, and only then will it progress to large-scale clinical trials in relatively healthy people to look for the prevention of disease. Most of us will choose to wait for this second stage of trials before undergoing any new treatments. Until then, prophylactic plasma transfusions will be the domain of the optimistic and the gullible.
DNA- and RNA-based electronic implants may revolutionize healthcare
The test tubes contain tiny DNA/enzyme-based circuits, which comprise TRUMPET, a new type of electronic device, smaller than a cell.
Implantable electronic devices can significantly improve patients’ quality of life. A pacemaker can encourage the heart to beat more regularly. A neural implant, usually placed at the back of the skull, can help brain function and encourage higher neural activity. Current research on neural implants finds them helpful to patients with Parkinson’s disease, vision loss, hearing loss, and other nerve damage problems. Several of these implants, such as Elon Musk’s Neuralink, have already been approved by the FDA for human use.
Yet, pacemakers, neural implants, and other such electronic devices are not without problems. They require constant electricity, limited through batteries that need replacements. They also cause scarring. “The problem with doing this with electronics is that scar tissue forms,” explains Kate Adamala, an assistant professor of cell biology at the University of Minnesota Twin Cities. “Anytime you have something hard interacting with something soft [like muscle, skin, or tissue], the soft thing will scar. That's why there are no long-term neural implants right now.” To overcome these challenges, scientists are turning to biocomputing processes that use organic materials like DNA and RNA. Other promised benefits include “diagnostics and possibly therapeutic action, operating as nanorobots in living organisms,” writes Evgeny Katz, a professor of bioelectronics at Clarkson University, in his book DNA- And RNA-Based Computing Systems.
While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output.
Adamala’s research focuses on developing such biocomputing systems using DNA, RNA, proteins, and lipids. Using these molecules in the biocomputing systems allows the latter to be biocompatible with the human body, resulting in a natural healing process. In a recent Nature Communications study, Adamala and her team created a new biocomputing platform called TRUMPET (Transcriptional RNA Universal Multi-Purpose GatE PlaTform) which acts like a DNA-powered computer chip. “These biological systems can heal if you design them correctly,” adds Adamala. “So you can imagine a computer that will eventually heal itself.”
The basics of biocomputing
Biocomputing and regular computing have many similarities. Like regular computing, biocomputing works by running information through a series of gates, usually logic gates. A logic gate works as a fork in the road for an electronic circuit. The input will travel one way or another, giving two different outputs. An example logic gate is the AND gate, which has two inputs (A and B) and two different results. If both A and B are 1, the AND gate output will be 1. If only A is 1 and B is 0, the output will be 0 and vice versa. If both A and B are 0, the result will be 0. While a computer gives these inputs in binary code or "bits," such as a 0 or 1, biocomputing uses DNA strands as inputs, whether double or single-stranded, and often uses fluorescent RNA as an output. In this case, the DNA enters the logic gate as a single or double strand.
If the DNA is double-stranded, the system “digests” the DNA or destroys it, which results in non-fluorescence or “0” output. Conversely, if the DNA is single-stranded, it won’t be digested and instead will be copied by several enzymes in the biocomputing system, resulting in fluorescent RNA or a “1” output. And the output for this type of binary system can be expanded beyond fluorescence or not. For example, a “1” output might be the production of the enzyme insulin, while a “0” may be that no insulin is produced. “This kind of synergy between biology and computation is the essence of biocomputing,” says Stephanie Forrest, a professor and the director of the Biodesign Center for Biocomputing, Security and Society at Arizona State University.
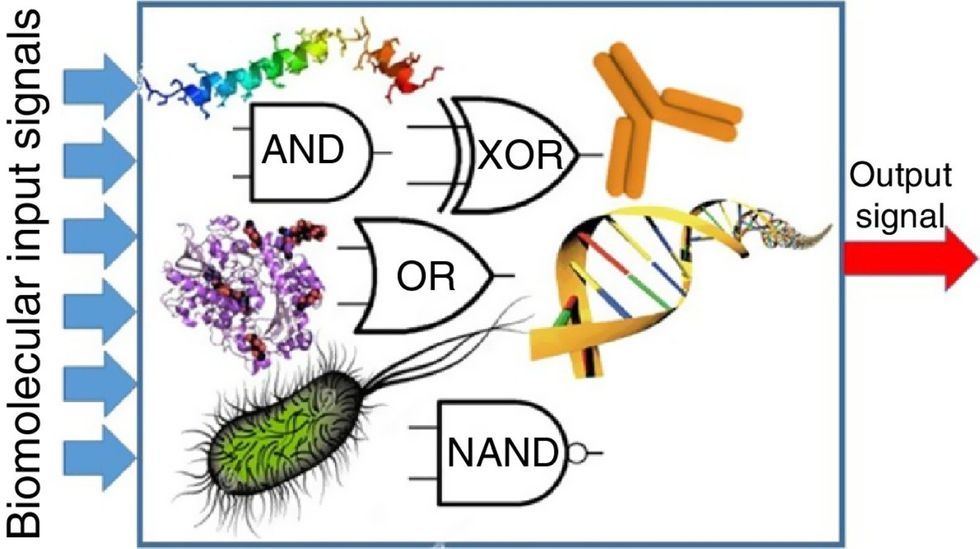
Biocomputing circles are made of DNA, RNA, proteins and even bacteria.
Evgeny Katz
The TRUMPET’s promise
Depending on whether the biocomputing system is placed directly inside a cell within the human body, or run in a test-tube, different environmental factors play a role. When an output is produced inside a cell, the cell's natural processes can amplify this output (for example, a specific protein or DNA strand), creating a solid signal. However, these cells can also be very leaky. “You want the cells to do the thing you ask them to do before they finish whatever their businesses, which is to grow, replicate, metabolize,” Adamala explains. “However, often the gate may be triggered without the right inputs, creating a false positive signal. So that's why natural logic gates are often leaky." While biocomputing outside a cell in a test tube can allow for tighter control over the logic gates, the outputs or signals cannot be amplified by a cell and are less potent.
TRUMPET, which is smaller than a cell, taps into both cellular and non-cellular biocomputing benefits. “At its core, it is a nonliving logic gate system,” Adamala states, “It's a DNA-based logic gate system. But because we use enzymes, and the readout is enzymatic [where an enzyme replicates the fluorescent RNA], we end up with signal amplification." This readout means that the output from the TRUMPET system, a fluorescent RNA strand, can be replicated by nearby enzymes in the platform, making the light signal stronger. "So it combines the best of both worlds,” Adamala adds.
These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body.
The TRUMPET biocomputing process is relatively straightforward. “If the DNA [input] shows up as single-stranded, it will not be digested [by the logic gate], and you get this nice fluorescent output as the RNA is made from the single-stranded DNA, and that's a 1,” Adamala explains. "And if the DNA input is double-stranded, it gets digested by the enzymes in the logic gate, and there is no RNA created from the DNA, so there is no fluorescence, and the output is 0." On the story's leading image above, if the tube is "lit" with a purple color, that is a binary 1 signal for computing. If it's "off" it is a 0.
While still in research, TRUMPET and other biocomputing systems promise significant benefits to personalized healthcare and medicine. These organic-based systems could detect cancer cells or low insulin levels inside a patient’s body. The study’s lead author and graduate student Judee Sharon is already beginning to research TRUMPET's ability for earlier cancer diagnoses. Because the inputs for TRUMPET are single or double-stranded DNA, any mutated or cancerous DNA could theoretically be detected from the platform through the biocomputing process. Theoretically, devices like TRUMPET could be used to detect cancer and other diseases earlier.
Adamala sees TRUMPET not only as a detection system but also as a potential cancer drug delivery system. “Ideally, you would like the drug only to turn on when it senses the presence of a cancer cell. And that's how we use the logic gates, which work in response to inputs like cancerous DNA. Then the output can be the production of a small molecule or the release of a small molecule that can then go and kill what needs killing, in this case, a cancer cell. So we would like to develop applications that use this technology to control the logic gate response of a drug’s delivery to a cell.”
Although platforms like TRUMPET are making progress, a lot more work must be done before they can be used commercially. “The process of translating mechanisms and architecture from biology to computing and vice versa is still an art rather than a science,” says Forrest. “It requires deep computer science and biology knowledge,” she adds. “Some people have compared interdisciplinary science to fusion restaurants—not all combinations are successful, but when they are, the results are remarkable.”
Crickets are low on fat, high on protein, and can be farmed sustainably. They are also crunchy.
In today’s podcast episode, Leaps.org Deputy Editor Lina Zeldovich speaks about the health and ecological benefits of farming crickets for human consumption with Bicky Nguyen, who joins Lina from Vietnam. Bicky and her business partner Nam Dang operate an insect farm named CricketOne. Motivated by the idea of sustainable and healthy protein production, they started their unconventional endeavor a few years ago, despite numerous naysayers who didn’t believe that humans would ever consider munching on bugs.
Yet, making creepy crawlers part of our diet offers many health and planetary advantages. Food production needs to match the rise in global population, estimated to reach 10 billion by 2050. One challenge is that some of our current practices are inefficient, polluting and wasteful. According to nonprofit EarthSave.org, it takes 2,500 gallons of water, 12 pounds of grain, 35 pounds of topsoil and the energy equivalent of one gallon of gasoline to produce one pound of feedlot beef, although exact statistics vary between sources.
Meanwhile, insects are easy to grow, high on protein and low on fat. When roasted with salt, they make crunchy snacks. When chopped up, they transform into delicious pâtes, says Bicky, who invents her own cricket recipes and serves them at industry and public events. Maybe that’s why some research predicts that edible insects market may grow to almost $10 billion by 2030. Tune in for a delectable chat on this alternative and sustainable protein.
Listen on Apple | Listen on Spotify | Listen on Stitcher | Listen on Amazon | Listen on Google
Further reading:
More info on Bicky Nguyen
https://yseali.fulbright.edu.vn/en/faculty/bicky-n...
The environmental footprint of beef production
https://www.earthsave.org/environment.htm
https://www.watercalculator.org/news/articles/beef-king-big-water-footprints/
https://www.frontiersin.org/articles/10.3389/fsufs.2019.00005/full
https://ourworldindata.org/carbon-footprint-food-methane
Insect farming as a source of sustainable protein
https://www.insectgourmet.com/insect-farming-growing-bugs-for-protein/
https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/insect-farming
Cricket flour is taking the world by storm
https://www.cricketflours.com/
https://talk-commerce.com/blog/what-brands-use-cricket-flour-and-why/

Lina Zeldovich has written about science, medicine and technology for Popular Science, Smithsonian, National Geographic, Scientific American, Reader’s Digest, the New York Times and other major national and international publications. A Columbia J-School alumna, she has won several awards for her stories, including the ASJA Crisis Coverage Award for Covid reporting, and has been a contributing editor at Nautilus Magazine. In 2021, Zeldovich released her first book, The Other Dark Matter, published by the University of Chicago Press, about the science and business of turning waste into wealth and health. You can find her on http://linazeldovich.com/ and @linazeldovich.